Inhibition of Cyclin-Dependent Kinase 5: A Strategy to Improve Sorafenib Response in Hepatocellular Carcinoma Therapy
- PMID: 30033593
- PMCID: PMC6590289
- DOI: 10.1002/hep.30190
Inhibition of Cyclin-Dependent Kinase 5: A Strategy to Improve Sorafenib Response in Hepatocellular Carcinoma Therapy
Abstract
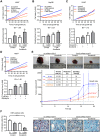
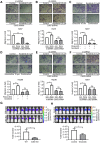
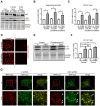
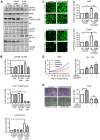
Therapeutic options for patients with advanced-stage hepatocellular carcinoma (HCC) are very limited. The only approved first-line treatment is the multi-tyrosine kinase inhibitor sorafenib, which shows low response rates and severe side effects. In particular, the compensatory activation of growth factor receptors leads to chemoresistance and limits the clinical impact of sorafenib. However, combination approaches to improve sorafenib have failed. Here we investigate the inhibition of cyclin-dependent kinase 5 (Cdk5) as a promising combination strategy to improve sorafenib response in HCC. Combination of sorafenib with Cdk5 inhibition (genetic knockdown by short hairpin RNA or CRISPR/Cas9 and pharmacologic inhibition) synergistically impaired HCC progression in vitro and in vivo by inhibiting both tumor cell proliferation and migration. Importantly, these effects were mediated by a mechanism for Cdk5: A liquid chromatography-tandem mass spectrometry-based proteomic approach revealed that Cdk5 inhibition interferes with intracellular trafficking, a process crucial for cellular homeostasis and growth factor receptor signaling. Cdk5 inhibition resulted in an accumulation of enlarged vesicles and respective cargos in the perinuclear region, considerably impairing the extent and quality of growth factor receptor signaling. Thereby, Cdk5 inhibition offers a comprehensive approach to globally disturb growth factor receptor signaling that is superior to specific inhibition of individual growth factor receptors. Conclusion: Cdk5 inhibition represents an effective approach to improve sorafenib response and to prevent sorafenib treatment escape in HCC. Notably, Cdk5 is an addressable target frequently overexpressed in HCC, and with Dinaciclib, a clinically tested Cdk5 inhibitor is readily available. Thus, our study provides evidence for clinically evaluating the combination of sorafenib and Dinaciclib to improve the therapeutic situation for patients with advanced-stage HCC.
© 2018 The Authors. Hepatology published by Wiley Periodicals, Inc., on behalf of the American Association for the Study of Liver Diseases.
Conflict of interest statement
Nothing to report.
Figures


References
-
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin 2011;61:69‐90. - PubMed
-
- Forner A, Gilabert M, Bruix J, Raoul JL. Treatment of intermediate‐stage hepatocellular carcinoma. Nat Rev Clin Oncol 2014;11:525‐535. - PubMed
-
- Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378‐390. - PubMed
-
- Nishida N, Kitano M, Sakurai T, Kudo M. Molecular mechanism and prediction of sorafenib chemoresistance in human hepatocellular carcinoma. Dig Dis 2015;33:771‐779. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

