Association Between Gut Microbiota and CD4 Recovery in HIV-1 Infected Patients
- PMID: 30034377
- PMCID: PMC6043814
- DOI: 10.3389/fmicb.2018.01451
Association Between Gut Microbiota and CD4 Recovery in HIV-1 Infected Patients
Abstract
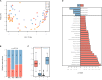
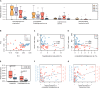
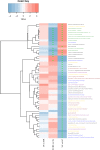
Composition of the gut microbiota has been linked with human immunedeficiency virus (HIV)-infected patients on antiretroviral therapy (ART). Evidence suggests that ART-treated patients with poor CD4+ T-cell recovery have higher levels of microbial translocation and immune activation. However, the association of the gut microbiota and immune recovery remains unclear. We performed a cross-sectional study on 30 healthy controls (HC) and 61 HIV-infected individuals, including 15 immunological ART responders (IRs), 20 immunological ART non-responders (INRs) and 26 untreated individuals (VU). IR and INR groups were classified by CD4+ T-cell counts of ≥350 cells/mm3 and <350 cells/mm3 after 2 years of ART, respectively. Each subject's gut microbiota composition was analyzed by metagenomics sequencing. Levels of CD4+ T cells, CD8+HLA-DR+ T cells and CD8+CD38+ T cells were measured by flow cytometry. We identified more Prevotella and fewer Bacteroides in HIV-infected individuals than in HC. Patients in INR group were enriched with Faecalibacterium prausnitzii, unclassified Subdoligranulum sp. and Coprococcus comes when compared with those in IR group. F. prausnitzii and unclassified Subdoligranulum sp. were overrepresented in individuals in VU group with CD4+ T-cell counts <350 cells/mm3. Moreover, we found that the relative abundance of unclassified Subdoligranulum sp. and C. comes were positively correlated with CD8+HLA-DR+ T-cell count and CD8+HLA-DR+/CD8+ percentage. Our study has shown that gut microbiota changes were associated with CD4+ T-cell counts and immune activation in HIV-infected subjects. Interventions to reverse gut dysbiosis and inhibit immune activation could be a new strategy for improving immune reconstitution of HIV-1-infected individuals.
Keywords: CD4 recovery; HIV-infected individuals; butyrate-producing bacteria; gut microbiota; metagenomics sequencing.
Figures
References
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate − a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57 289–300.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

