Metallacarboranes as tunable redox potential electrochemical indicators for screening of gene mutation
- PMID: 30034717
- PMCID: PMC6022041
- DOI: 10.1039/c6sc01567k
Metallacarboranes as tunable redox potential electrochemical indicators for screening of gene mutation
Abstract
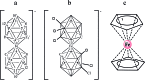
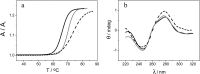
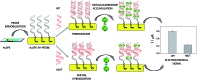
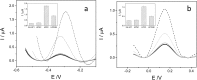
The substitution of hydrogen with chlorine in the metallacarborane [3,3-Fe(1,2-closo-C2B9H11)2]- cluster modulates the formal potential of the Fe3+/Fe2+ redox couple, shifting it to a more positive value. Hence, very similar redox probes with a wide range of formal potentials, ranging from negative to positive values, are available. Thus, we have achieved the synthesis and studied the electrochemical behaviour of the sodium salt of [3,3-Fe(8,9,12-Cl3-1,2-closo-C2B9H8)2]- in aqueous media. This strategy allows tuning of the redox potential of the [3,3-Fe(1,2-closo-C2B9H11)2]- framework with a minor change in its shape and dimensions. We also describe the interaction of the prepared [3,3-Fe(8,9,12-Cl3-1,2-closo-C2B9H8)2]- and the pristine [3,3-Fe(1,2-closo-C2B9H11)2]- with DNA. These studies have been carried out not only with DNA in solution but also with DNA immobilized on screen-printed gold electrodes. The results obtained point to a strong interaction between the metallacarboranes and DNA, to a different extent with single stranded DNA (ssDNA) compared to double stranded DNA (dsDNA). This property makes them selective and wide-ranging potential electrochemical indicators of hybridization. The suitability of these new redox indicators for selective DNA biosensor development has been probed by the direct detection of two different mutations associated with cystic fibrosis in PCR amplicons extracted from blood cells.
Figures





References
-
- Palaska P., Aritzoglou E., Girousi S. Talanta. 2007;72:1119–1206. - PubMed
-
- Erdem A. Talanta. 2007;74:318–325. - PubMed
-
- Wang J. Trends Anal. Chem. 2002;21:226–232.
-
- Shabani A., Mak A. W. H., Gerges I., Cuccia L. A., Lawrence M. F. Talanta. 2006;70:615–623. - PubMed
-
- Lin X. H., Wu P., Chen W., Zhang Y. F., Xia X. H. Talanta. 2007;72:468–471. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

