Gating mechanisms during actin filament elongation by formins
- PMID: 30035712
- PMCID: PMC6056239
- DOI: 10.7554/eLife.37342
Gating mechanisms during actin filament elongation by formins
Abstract
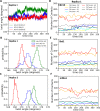
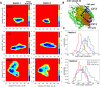
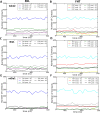
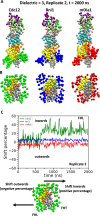
Formins play an important role in the polymerization of unbranched actin filaments, and particular formins slow elongation by 5-95%. We studied the interactions between actin and the FH2 domains of formins Cdc12, Bni1 and mDia1 to understand the factors underlying their different rates of polymerization. All-atom molecular dynamics simulations revealed two factors that influence actin filament elongation and correlate with the rates of elongation. First, FH2 domains can sterically block the addition of new actin subunits. Second, FH2 domains flatten the helical twist of the terminal actin subunits, making the end less favorable for subunit addition. Coarse-grained simulations over longer time scales support these conclusions. The simulations show that filaments spend time in states that either allow or block elongation. The rate of elongation is a time-average of the degree to which the formin compromises subunit addition rather than the formin-actin complex literally being in 'open' or 'closed' states.
Keywords: FH2 domain; S. cerevisiae; S. pombe; coarse-grained; formin; gating mechanism; intermolecular interactions; molecular biophysics; molecular dynamics; mouse; structural biology.
© 2018, Aydin et al.
Conflict of interest statement
FA, NC, TP, GV No competing interests declared
Figures
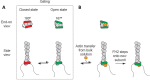

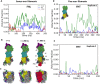
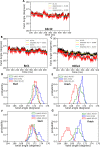
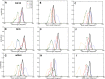




References
-
- Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. Journal of Computational Chemistry. 1983;4:187–217. doi: 10.1002/jcc.540040211. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

