Haemophilus parasuis α-2,3-sialyltransferase-mediated lipooligosaccharide sialylation contributes to bacterial pathogenicity
- PMID: 30036124
- PMCID: PMC6104685
- DOI: 10.1080/21505594.2018.1502606
Haemophilus parasuis α-2,3-sialyltransferase-mediated lipooligosaccharide sialylation contributes to bacterial pathogenicity
Abstract
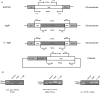
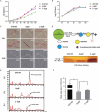
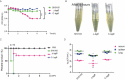
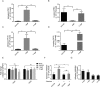
Bacterial lipooligosaccharide (LOS) is an important virulence-associated factor, and its sialylation largely confers its ability to mediate cell adhesion, invasion, inflammation, and immune evasion. Here, we investigated the function of the Haemophilus parasuis α-2,3-sialyltransferase gene, lsgB, which determines the terminal sialylation of LOS, by generating a lsgB deletion mutant as well as a complementation strain. Our data indicate a direct effect of lsgB on LOS sialylation and reveal important roles of lsgB in promoting the pathogenicity of H. parasuis, including adhesion to and invasion of porcine cells in vitro, bacterial load and survival in vivo, as well as a contribution to serum resistance. These observations highlight the function of lsgB in mediating LOS sialylation and more importantly its role in H. parasuis infection. These findings provide a more profound understanding of the pathogenic mechanism of this disease-causing bacterium.
Keywords: 3-sialyltransferase; LOS sialylation; adhesion and invasion; lsgB; pathogenicity; serum resistance; α-2.
Figures

Similar articles
-
Distribution of genes involved in sialic acid utilization in strains of Haemophilus parasuis.Microbiology (Reading). 2012 Aug;158(Pt 8):2117-2124. doi: 10.1099/mic.0.056994-0. Epub 2012 May 18. Microbiology (Reading). 2012. PMID: 22609756
-
Deletion of the rfaE gene in Haemophilus parasuis SC096 strain attenuates serum resistance, adhesion and invasion.Microb Pathog. 2014 Sep;74:33-7. doi: 10.1016/j.micpath.2014.07.006. Epub 2014 Jul 28. Microb Pathog. 2014. PMID: 25078003
-
Sialylated Lipooligosaccharide Contributes to Glaesserella parasuis Penetration of Porcine Respiratory Epithelial Barrier.ACS Infect Dis. 2021 Mar 12;7(3):661-671. doi: 10.1021/acsinfecdis.0c00850. Epub 2021 Mar 1. ACS Infect Dis. 2021. PMID: 33645216
-
Advances in the quest for virulence factors of Haemophilus parasuis.Vet J. 2013 Dec;198(3):571-6. doi: 10.1016/j.tvjl.2013.08.027. Epub 2013 Sep 4. Vet J. 2013. PMID: 24084037 Review.
-
Update on the pathogenesis of Haemophilus parasuis infection and virulence factors.Vet Microbiol. 2014 Jan 10;168(1):1-7. doi: 10.1016/j.vetmic.2013.07.027. Epub 2013 Aug 12. Vet Microbiol. 2014. PMID: 23972951 Review.
Cited by
-
Glycosphingolipids: from metabolism to chemoenzymatic total synthesis.Org Biomol Chem. 2024 Aug 22;22(33):6665-6683. doi: 10.1039/d4ob00695j. Org Biomol Chem. 2024. PMID: 39120686 Free PMC article. Review.
-
Molecular characterization of Glaesserella parasuis strains circulating in North American swine production systems.BMC Vet Res. 2023 Aug 28;19(1):135. doi: 10.1186/s12917-023-03698-x. BMC Vet Res. 2023. PMID: 37641044 Free PMC article.
-
Application of mouse model for evaluation of recombinant LpxC and GmhA as novel antigenic vaccine candidates of Glaesserella parasuis serotype 13.J Vet Med Sci. 2021 Oct 2;83(10):1500-1508. doi: 10.1292/jvms.21-0298. Epub 2021 Aug 16. J Vet Med Sci. 2021. PMID: 34393140 Free PMC article.
-
Genomic insight into the diversity of Glaesserella parasuis isolates from 19 countries.mSphere. 2024 Sep 25;9(9):e0023124. doi: 10.1128/msphere.00231-24. Epub 2024 Aug 28. mSphere. 2024. PMID: 39194201 Free PMC article.
-
Epidemiology and pathogenicity of Haemophilus parasuis in eastern China.Front Microbiol. 2025 May 15;16:1589975. doi: 10.3389/fmicb.2025.1589975. eCollection 2025. Front Microbiol. 2025. PMID: 40443997 Free PMC article.
References
-
- Oliveira S, Pijoan C.. Haemophilus parasuis: new trends on diagnosis, epidemiology and control. Vet Microbiol. 2004;99:1–12. - PubMed
-
- Huang J, Wang X, Cao Q, et al. ClpP participates in stress tolerance and negatively regulates biofilm formation in Haemophilus parasuis. Vet Microbiol. 2016;182:141–149. - PubMed
-
- Cai X, Chen H, Blackall PJ, et al. Serological characterization of Haemophilus parasuis isolates from China. Vet Microbiol. 2005;111:231–236. - PubMed
-
- Rubies X, Kielstein P, Costa L, et al. Prevalence of Haemophilus parasuis serovars isolated in Spain from 1993 to 1997. Vet Microbiol. 1999;66:245–248. - PubMed
-
- Lichtensteiger CA, Vimr ER. Purification and renaturation of membrane neuraminidase from Haemophilus parasuis. Vet Microbiol. 2003;93:79–87. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
