Physiological Starvation Promotes Caenorhabditis elegans Vulval Induction
- PMID: 30037804
- PMCID: PMC6118308
- DOI: 10.1534/g3.118.200449
Physiological Starvation Promotes Caenorhabditis elegans Vulval Induction
Abstract
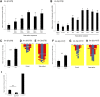
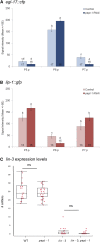
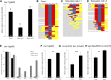
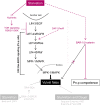
Studying how molecular pathways respond to ecologically relevant environmental variation is fundamental to understand organismal development and its evolution. Here we characterize how starvation modulates Caenorhabditis elegans vulval cell fate patterning - an environmentally sensitive process, with a nevertheless robust output. Past research has shown many vulval mutants affecting EGF-Ras-MAPK, Delta-Notch and Wnt pathways to be suppressed by environmental factors, such as starvation. Here we aimed to resolve previous, seemingly contradictory, observations on how starvation modulates levels of vulval induction. Using the strong starvation suppression of the Vulvaless phenotype of lin-3/egf reduction-of-function mutations as an experimental paradigm, we first tested for a possible involvement of the sensory system in relaying starvation signals to affect vulval induction: mutation of various sensory inputs, DAF-2/Insulin or DAF-7/TGF-β signaling did not abolish lin-3(rf) starvation suppression. In contrast, nutrient deprivation induced by mutation of the intestinal peptide transporter gene pept-1 or the TOR pathway component rsks-1 (the ortholog of mammalian P70S6K) very strongly suppressed lin-3(rf) mutant phenotypes. Therefore, physiologically starved animals induced by these mutations tightly recapitulated the effects of external starvation on vulval induction. While both starvation and pept-1 RNAi were sufficient to increase Ras and Notch pathway activities in vulval cells, the highly penetrant Vulvaless phenotype of a tissue-specific null allele of lin-3 was not suppressed by either condition. This and additional results indicate that partial lin-3 expression is required for starvation to affect vulval induction. These results suggest a cross-talk between nutrient deprivation, TOR-S6K and EGF-Ras-MAPK signaling during C. elegans vulval induction.
Keywords: Caenorhabditis; LIN-3; TOR-S6K; mutational penetrance; vulval cell fate patterning.
Copyright © 2018 Grimbert et al.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
