A GPI Signal Peptide-Anchored Split-Ubiquitin (GPS) System for Detecting Soluble Bait Protein Interactions at the Membrane
- PMID: 30037807
- PMCID: PMC6130019
- DOI: 10.1104/pp.18.00577
A GPI Signal Peptide-Anchored Split-Ubiquitin (GPS) System for Detecting Soluble Bait Protein Interactions at the Membrane
Abstract
Bait fusion proteins with a glycosyl-phosphatidylinositol signal sequence anchor enable effective split ubiquitin screening for interactions with otherwise soluble membrane proteins.
Figures
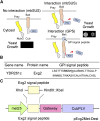
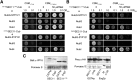
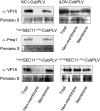
Comment in
-
An Improved Tool for Mapping the Membrane-Associated Protein Interactome.Plant Physiol. 2018 Sep;178(1):11-12. doi: 10.1104/pp.18.00969. Plant Physiol. 2018. PMID: 30194264 Free PMC article. No abstract available.
References
-
- Caro LHP, Tettelin H, Vossen JH, Ram AF, van den Ende H, Klis FM (1997) In silicio identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 13: 1477–1489 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/I024496/1/Biotechnology and Biological Sciences Research Council /International
- BB/K015893/1/Biotechnology and Biological Sciences Research Council /International
- BB/L001276/1/Biotechnology and Biological Sciences Research Council /International
- BB/M01133X/1/Biotechnology and Biological Sciences Research Council /International
- BB/M001601/1/Biotechnology and Biological Sciences Research Council /International
LinkOut - more resources
Full Text Sources
Other Literature Sources

