Molecular Portrait of Hypoxia in Breast Cancer: A Prognostic Signature and Novel HIF-Regulated Genes
- PMID: 30037853
- PMCID: PMC6279594
- DOI: 10.1158/1541-7786.MCR-18-0345
Molecular Portrait of Hypoxia in Breast Cancer: A Prognostic Signature and Novel HIF-Regulated Genes
Abstract
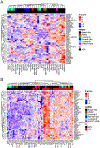
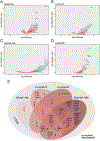
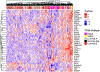
Intratumoral hypoxia has been associated with invasion, metastasis, and treatment failure, prompting the need for a global characterization of the response to hypoxic conditions. The current study presents the results of a large-scale RNA sequencing (RNA-seq) effort, analyzing 31 breast cancer cell lines representative of breast cancer subtypes or normal mammary epithelial (NME) cells exposed to control tissue culture conditions (20% O2) or hypoxic conditions (1% O2). The results demonstrate that NME have a stronger response to hypoxia both in terms of number of genes induced by hypoxia as well as level of expression. A conserved 42-gene hypoxia signature shared across PAM50 subtypes and genes that are exclusively upregulated in Luminal A, Luminal B, and normal-like mammary epithelial cells is identified. The 42-gene expression signature is enriched in a subset of basal-like cell lines and tumors and differentiates survival among patients with basal-like tumors. Mechanistically, the hypoxia-inducible factors (HIF-1 and/or HIF-2) mediate the conserved hypoxic response. Also, four novel hypoxia-regulated and HIF-1-responsive genes were identified as part of the conserved signature. This dataset provides a novel resource to query transcriptional changes that occur in response to hypoxia and serves as a starting point for a clinical assay to aid in stratifying patients that would benefit from hypoxia-targeted therapies, some of which are currently in clinical trials. IMPLICATIONS: RNA-seq of 31 breast cancer cells exposed to control or hypoxic conditions reveals a conserved genomic signature that contains novel HIF-regulated genes and is prognostic for the survival of patients with triple-negative breast cancer.
©2018 American Association for Cancer Research.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Dewhirst MW. Intermittent hypoxia furthers the rationale for hypoxia-inducible factor-1 targeting. Cancer Res 2007;67(3):854–5. - PubMed
-
- Brahimi-Horn MC, Chiche J, Pouyssegur J. Hypoxia and cancer. J Mol Med 2007;85(12):1301–7. - PubMed
-
- Semenza GL, Ruvolo PP. Introduction to tumor microenvironment regulation of cancer cell survival, metastasis, inflammation, and immune surveillance. Biochim Biophys Acta 2016;1863(3):379–81. - PubMed
-
- Gillies RJ, Gatenby RA. Hypoxia and adaptive landscapes in the evolution of carcinogenesis. Cancer Metastasis Rev 2007;26(2):311–7. - PubMed
-
- Bos R, van der Groep P, Greijer AE, Shvarts A, Meijer S, Pinedo HM, et al. Levels of hypoxia-inducible factor-1α independently predict prognosis in patients with lymph node negative breast carcinoma. Cancer 2003;97(6):1573–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

