Ribosome Profiling: Global Views of Translation
- PMID: 30037969
- PMCID: PMC6496350
- DOI: 10.1101/cshperspect.a032698
Ribosome Profiling: Global Views of Translation
Abstract
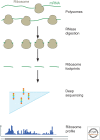
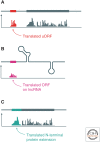
The translation of messenger RNA (mRNA) into protein and the folding of the resulting protein into an active form are prerequisites for virtually every cellular process and represent the single largest investment of energy by cells. Ribosome profiling-based approaches have revolutionized our ability to monitor every step of protein synthesis in vivo, allowing one to measure the rate of protein synthesis across the proteome, annotate the protein coding capacity of genomes, monitor localized protein synthesis, and explore cotranslational folding and targeting. The rich and quantitative nature of ribosome profiling data provides an unprecedented opportunity to explore and model complex cellular processes. New analytical techniques and improved experimental protocols will provide a deeper understanding of the factors controlling translation speed and its impact on protein function and cell physiology as well as the role of ribosomal RNA and mRNA modifications in regulating translation.
Copyright © 2019 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures



References
-
- Alain T, Morita M, Fonseca BD, Yanagiya A, Siddiqui N, Bhat M, Zammit D, Marcus V, Metrakos P, Voyer LA, et al. 2012. eIF4E/4E-BP ratio predicts the efficacy of mTOR targeted therapies. Cancer Res 72: 6468–6476. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
