RNA triphosphatase DUSP11 enables exonuclease XRN-mediated restriction of hepatitis C virus
- PMID: 30038017
- PMCID: PMC6094126
- DOI: 10.1073/pnas.1802326115
RNA triphosphatase DUSP11 enables exonuclease XRN-mediated restriction of hepatitis C virus
Abstract
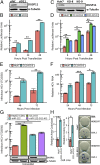
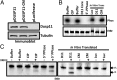
Seventy percent of people infected with hepatitis C virus (HCV) will suffer chronic infection, putting them at risk for liver disease, including hepatocellular carcinoma. The full range of mechanisms that render some people more susceptible to chronic infection and liver disease is still being elucidated. XRN exonucleases can restrict HCV replication and may help to resolve HCV infections. However, it is unknown how 5' triphosphorylated HCV transcripts, primary products of the viral polymerase, become susceptible to attack by 5' monophosphate-specific XRNs. Here, we show that the 5' RNA triphosphatase DUSP11 acts on HCV transcripts, rendering them susceptible to XRN-mediated attack. Cells lacking DUSP11 show substantially enhanced HCV replication, and this effect is diminished when XRN expression is reduced. MicroRNA-122 (miR-122), a target of current phase II anti-HCV drugs, is known to protect HCV transcripts against XRNs. We show that HCV replication is less dependent on miR-122 in cells lacking DUSP11. Combined, these results implicate DUSP11 as an important component of XRN-mediated restriction of HCV.
Keywords: miR-122; microRNA; restriction factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Kanwal F, et al. Risk of hepatocellular cancer in HCV patients treated with direct-acting antiviral agents. Gastroenterology. 2017;153:996–1005.e1. - PubMed
-
- Missiha SB, Ostrowski M, Heathcote EJ. Disease progression in chronic hepatitis C: Modifiable and nonmodifiable factors. Gastroenterology. 2008;134:1699–1714. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

