Cell-specific histone modification maps in the human frontal lobe link schizophrenia risk to the neuronal epigenome
- PMID: 30038276
- PMCID: PMC6063773
- DOI: 10.1038/s41593-018-0187-0
Cell-specific histone modification maps in the human frontal lobe link schizophrenia risk to the neuronal epigenome
Abstract
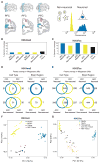
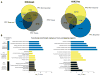
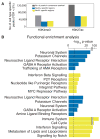
Risk variants for schizophrenia affect more than 100 genomic loci, yet cell- and tissue-specific roles underlying disease liability remain poorly characterized. We have generated for two cortical areas implicated in psychosis, the dorsolateral prefrontal cortex and anterior cingulate cortex, 157 reference maps from neuronal, neuron-depleted and bulk tissue chromatin for two histone marks associated with active promoters and enhancers, H3-trimethyl-Lys4 (H3K4me3) and H3-acetyl-Lys27 (H3K27ac). Differences between neuronal and neuron-depleted chromatin states were the major axis of variation in histone modification profiles, followed by substantial variability across subjects and cortical areas. Thousands of significant histone quantitative trait loci were identified in neuronal and neuron-depleted samples. Risk variants for schizophrenia, depressive symptoms and neuroticism were significantly over-represented in neuronal H3K4me3 and H3K27ac landscapes. Our Resource, sponsored by PsychENCODE and CommonMind, highlights the critical role of cell-type-specific signatures at regulatory and disease-associated noncoding sequences in the human frontal lobe.
Conflict of interest statement
Figures



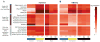
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 MH094714/MH/NIMH NIH HHS/United States
- U01 MH103365/MH/NIMH NIH HHS/United States
- U01 AG016976/AG/NIA NIH HHS/United States
- U01 MH103392/MH/NIMH NIH HHS/United States
- R01 MH109677/MH/NIMH NIH HHS/United States
- R01 HL105756/HL/NHLBI NIH HHS/United States
- U01 MH103346/MH/NIMH NIH HHS/United States
- R01 AG033193/AG/NIA NIH HHS/United States
- U01 MH103340/MH/NIMH NIH HHS/United States
- P50 MH084053/MH/NIMH NIH HHS/United States
- P50 MH066392/MH/NIMH NIH HHS/United States
- R01 MH080405/MH/NIMH NIH HHS/United States
- R01 MH085542/MH/NIMH NIH HHS/United States
- P50 MH106934/MH/NIMH NIH HHS/United States
- R01 AG050986/AG/NIA NIH HHS/United States
- R37 MH057881/MH/NIMH NIH HHS/United States
- U24 AG021886/AG/NIA NIH HHS/United States
- R21 MH103877/MH/NIMH NIH HHS/United States
- R01 MH075916/MH/NIMH NIH HHS/United States
- U01 AG032984/AG/NIA NIH HHS/United States
- R01 MH105472/MH/NIMH NIH HHS/United States
- R01 MH106056/MH/NIMH NIH HHS/United States
- U01 MH103339/MH/NIMH NIH HHS/United States
- I01 BX002395/BX/BLRD VA/United States
- R01 MH097276/MH/NIMH NIH HHS/United States
- P01 AG002219/AG/NIA NIH HHS/United States
- R01 MH093725/MH/NIMH NIH HHS/United States
- P50 AG005138/AG/NIA NIH HHS/United States
- R21 MH102791/MH/NIMH NIH HHS/United States
- R01 MH105898/MH/NIMH NIH HHS/United States
- R21 MH105881/MH/NIMH NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

