Murine colitis reveals a disease-associated bacteriophage community
- PMID: 30038310
- PMCID: PMC6112176
- DOI: 10.1038/s41564-018-0210-y
Murine colitis reveals a disease-associated bacteriophage community
Abstract
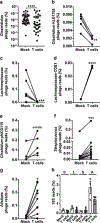
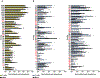
The dysregulation of intestinal microbial communities is associated with inflammatory bowel diseases (IBD). Studies aimed at understanding the contribution of the microbiota to inflammatory diseases have primarily focused on bacteria, yet the intestine harbours a viral component dominated by prokaryotic viruses known as bacteriophages (phages). Phage numbers are elevated at the intestinal mucosal surface and phages increase in abundance during IBD, suggesting that phages play an unidentified role in IBD. We used a sequence-independent approach for the selection of viral contigs and then applied quantitative metagenomics to study intestinal phages in a mouse model of colitis. We discovered that during colitis the intestinal phage population is altered and transitions from an ordered state to a stochastic dysbiosis. We identified phages specific to pathobiotic hosts associated with intestinal disease, whose abundances are altered during colitis. Additionally, phage populations in healthy and diseased mice overlapped with phages from healthy humans and humans with IBD. Our findings indicate that intestinal phage communities are altered during inflammatory disease, establishing a platform for investigating phage involvement in IBD.
Conflict of interest statement
Competing interests
The authors declare no competing interests related to this work.
Figures


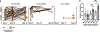
Comment in
-
Bacteriophage virome in IBD.Nat Rev Gastroenterol Hepatol. 2018 Sep;15(9):520. doi: 10.1038/s41575-018-0056-z. Nat Rev Gastroenterol Hepatol. 2018. PMID: 30076371 No abstract available.
References
-
- Molodecky NA et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 142, 46–54 e42 (2012). - PubMed
-
- Hooper LV, Midtvedt T & Gordon JI How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr 22, 283–307 (2002). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

