Dynamic interplay between enhancer-promoter topology and gene activity
- PMID: 30038397
- PMCID: PMC6119122
- DOI: 10.1038/s41588-018-0175-z
Dynamic interplay between enhancer-promoter topology and gene activity
Abstract
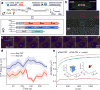
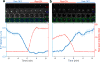
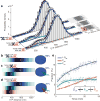
A long-standing question in gene regulation is how remote enhancers communicate with their target promoters, and specifically how chromatin topology dynamically relates to gene activation. Here, we combine genome editing and multi-color live imaging to simultaneously visualize physical enhancer-promoter interaction and transcription at the single-cell level in Drosophila embryos. By examining transcriptional activation of a reporter by the endogenous even-skipped enhancers, which are located 150 kb away, we identify three distinct topological conformation states and measure their transition kinetics. We show that sustained proximity of the enhancer to its target is required for activation. Transcription in turn affects the three-dimensional topology as it enhances the temporal stability of the proximal conformation and is associated with further spatial compaction. Furthermore, the facilitated long-range activation results in transcriptional competition at the locus, causing corresponding developmental defects. Our approach offers quantitative insight into the spatial and temporal determinants of long-range gene regulation and their implications for cellular fates.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Benoist C, Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981;290:304–10. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

