TRAF3IP2, a novel therapeutic target in glioblastoma multiforme
- PMID: 30038719
- PMCID: PMC6049871
- DOI: 10.18632/oncotarget.25710
TRAF3IP2, a novel therapeutic target in glioblastoma multiforme
Abstract
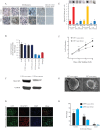
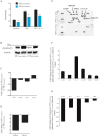
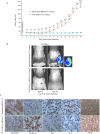
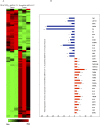
Glioblastoma multiforme (glioblastoma) remains one of the deadliest cancers. Pro-inflammatory and pro-tumorigenic mediators present in tumor microenvironment (TME) facilitate communication between tumor cells and adjacent non-malignant cells, resulting in glioblastoma growth. Since a majority of these mediators are products of NF-κB- and/or AP-1-responsive genes, and as TRAF3 Interacting Protein 2 (TRAF3IP2) is an upstream regulator of both transcription factors, we hypothesized that targeting TRAF3IP2 blunts tumor growth by inhibiting NF-κB and pro-inflammatory/pro-tumorigenic mediators. Our in vitro data demonstrate that similar to primary glioblastoma tumor tissues, malignant glioblastoma cell lines (U87 and U118) express high levels of TRAF3IP2. Silencing TRAF3IP2 expression inhibits basal and inducible NF-κB activation, induction of pro-inflammatory mediators, clusters of genes involved in cell cycle progression and angiogenesis, and formation of spheroids. Additionally, silencing TRAF3IP2 significantly increases apoptosis. In vivo studies indicate TRAF3IP2-silenced U87 cells formed smaller tumors. Additionally, treating existing tumors formed by wild type U87 cells with lentiviral TRAF3IP2 shRNA markedly regresses their size. Analysis of residual tumors revealed reduced expression of pro-inflammatory/pro-tumorigenic/pro-angiogenic mediators and kinesins. In contrast, the expression of IL-10, an anti-inflammatory cytokine, was increased. Together, these novel data indicate that TRAF3IP2 is a master regulator of malignant signaling in glioblastoma, and its targeting modulates the TME and inhibits tumor growth by suppressing the expression of mediators involved in inflammation, angiogenesis, growth, and malignant transformation. Our data identify TRAF3IP2 as a potential therapeutic target in glioblastoma growth and dissemination.
Keywords: TRAF3IP2; cancer stem cells; glioblastoma multiforme; inflammation; tumor microenvironment.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare no competing interests.
Figures


References
-
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. - DOI - PubMed
-
- Jotzu C, Alt E, Welte G, Li J, Hennessy BT, Devarajan E, Krishnappa S, Pinilla S, Droll L, Song YH. Adipose tissue-derived stem cells differentiate into carcinoma-associated fibroblast-like cells under the influence of tumor-derived factors. Anal Cell Pathol (Amst) 2010;33:61–79. doi: 10.3233/ACP-CLO-2010-0535. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

