Virus classification - where do you draw the line?
- PMID: 30039318
- PMCID: PMC6096723
- DOI: 10.1007/s00705-018-3938-z
Virus classification - where do you draw the line?
Abstract
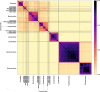
High-throughput sequencing (HTS) and its use in recovering and assembling novel virus sequences from environmental, human clinical, veterinary and plant samples has unearthed a vast new catalogue of viruses. Their classification, known by their sequences alone, sets a major challenge to traditional virus taxonomy, especially at the family and species levels, which have been historically based largely on descriptive taxon definitions. These typically entail some knowledge of their phenotypic properties, including replication strategies, virion structure and clinical and epidemiological features, such as host range, geographical distribution and disease outcomes. Little to no information on these attributes is available, however, for viruses identified in metagenomic datasets. If such viruses are to be included in virus taxonomy, their assignments will have to be guided largely or entirely by metrics of genetic relatedness. The immediate problem here is that the International Committee on Taxonomy of Viruses (ICTV), an organisation that authorises the taxonomic classification of viruses, provides little or no guidance on how similar or how divergent viruses must be in order to be considered members of new species or new families. We have recently developed a method for scoring genomic (dis)similarity between viruses (Genome Relationships Applied to Virus Taxonomy - GRAViTy) among the eukaryotic and prokaryotic viruses currently classified by the ICTV. At the family and genus levels, we found large-scale consistency between genetic relationships and their taxonomic assignments for eukaryotic viruses of all genome configurations and genome sizes. Family assignments of prokaryotic viruses have, however, been made at a quite different genetic level, and groupings currently classified as sub-families are a much better match to the eukaryotic virus family level. These findings support the ongoing reorganisation of bacteriophage taxonomy by the ICTV Phage Study Group. A rapid and objective means to explore metagenomic viral diversity and make evidence-based assignments for such viruses at each taxonomic layer is essential. Analysis of sequences by GRAViTy provides evidence that family (and genus) assignments of currently classified viruses are largely underpinned by genomic relatedness, and these features could serve as a guide towards an evidence-based classification of metagenomic viruses in the future.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Adams MJ, Lefkowitz EJ, King AM, Harrach B, Harrison RL, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Mushegian AR, Nibert ML, Sabanadzovic S, Sanfacon H, Siddell SG, Simmonds P, Varsani A, Zerbini FM, Orton RJ, Smith DB, Gorbalenya AE, Davison AJ. 50 years of the International Committee on Taxonomy of Viruses: progress and prospects. Arch Virol. 2017;162:1441–1446. doi: 10.1007/s00705-016-3215-y. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

