The dNTP triphosphohydrolase activity of SAMHD1 persists during S-phase when the enzyme is phosphorylated at T592
- PMID: 30039733
- PMCID: PMC6110608
- DOI: 10.1080/15384101.2018.1480216
The dNTP triphosphohydrolase activity of SAMHD1 persists during S-phase when the enzyme is phosphorylated at T592
Abstract
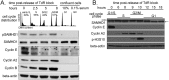
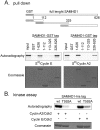
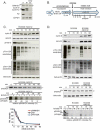
SAMHD1 is the major catabolic enzyme regulating the intracellular concentrations of DNA precursors (dNTPs). The S-phase kinase CDK2-cyclinA phosphorylates SAMHD1 at Thr-592. How this modification affects SAMHD1 function is highly debated. We investigated the role of endogenous SAMHD1 phosphorylation during the cell cycle. Thr-592 phosphorylation occurs first at the G1/S border and is removed during mitotic exit parallel with Thr-phosphorylations of most CDK1 targets. Differential sensitivity to the phosphatase inhibitor okadaic acid suggested different involvement of the PP1 and PP2 families dependent upon the time of the cell cycle. SAMHD1 turn-over indicates that Thr-592 phosphorylation does not cause rapid protein degradation. Furthermore, SAMHD1 influenced the size of the four dNTP pools independently of its phosphorylation. Our findings reveal that SAMHD1 is active during the entire cell cycle and performs an important regulatory role during S-phase by contributing with ribonucleotide reductase to maintain dNTP pool balance for proper DNA replication.
Keywords: Deoxynucleotide metabolism; Sterile alpha motif and HD domain containing protein1 (SAMHD1); cell cycle.
Figures




References
-
- Ji X, Tang C, Zhao Q, et al. Structural basis of cellular dNTP regulation by SAMHD1. Proceedings of the National Academy of Sciences [Internet]; 2014; 111:E4305–14. Available from: http://www.pnas.org/cgi/doi/10.1073/pnas.1412289111 - DOI - PMC - PubMed
-
- Tüngler V, Staroske W, Kind B, et al. Single-stranded nucleic acids promote SAMHD1 complex formation. J Mol Med. 2013;91:759–770. - PubMed
-
- Ryoo J, Choi J, Oh C, et al. The ribonuclease activity of SAMHD1 is required for HIV-1 restriction. Nat Med [Internet]. 2014;20:936–941. Available from: http://www.nature.com/doifinder/10.1038/nm.3626 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
