Engineering cell heterogeneity into organs-on-a-chip
- PMID: 30040104
- PMCID: PMC6081245
- DOI: 10.1039/c8lc00413g
Engineering cell heterogeneity into organs-on-a-chip
Abstract
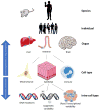
Organ-on-a-chip development is an application that will benefit from advances in cell heterogeneity characterization because these culture models are intended to mimic in vivo microenvironments, which are complex and dynamic. Due in no small part to advances in microfluidic single cell analysis methods, cell-to-cell variability is an increasingly understood feature of physiological tissues, with cell types from as common as 1 out of every 2 cells to as rare as 1 out of every 100 000 cells having important roles in the biochemical and biological makeup of tissues and organs. Variability between neighboring cells can be transient or maintained, and ordered or stochastic. This review covers three areas of well-studied cell heterogeneity that are informative for organ-on-a-chip development efforts: tumors, the lung, and the intestine. Then we look at how recent single cell analysis strategies have enabled better understanding of heterogeneity within in vitro and in vivo tissues. Finally, we provide a few work-arounds for adapting current on-chip culture methods to better mimic physiological cell heterogeneity including accounting for crucial rare cell types and events.
Conflict of interest statement
9. Conflicts of interest
There are no conflicts to declare.
Figures



References
-
- [accessed 3 April 2018];Growing human Organs-on-a-Chip. https://www.roche.com/research_and_development/what_we_are_working_on/re....
-
- Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, Li N, Szpankowski L, Fowler B, Chen P, Ramalingam N, Sun G, Thu M, Norris M, Lebofsky R, Toppani D, Kemp DW, Wong M, Clerkson B, Jones BN, Wu S, Knutsson L, Alvarado B, Wang J, Weaver LS, May AP, Jones RC, Unger MA, Kriegstein AR, West JAA. Nat Biotechnol. 2014;32:1053–1058. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

