Chitinase: diversity, limitations, and trends in engineering for suitable applications
- PMID: 30042170
- PMCID: PMC6131217
- DOI: 10.1042/BSR20180323
Chitinase: diversity, limitations, and trends in engineering for suitable applications
Abstract
Chitinases catalyze the degradation of chitin, a ubiquitous polymer generated from the cell walls of fungi, shells of crustaceans, and cuticles of insects. They are gaining increasing attention in medicine, agriculture, food and drug industries, and environmental management. Their roles in the degradation of chitin for the production of industrially useful products and in the control of fungal pathogens and insect pests render them attractive for such purposes. However, chitinases have diverse sources, characteristics, and mechanisms of action that seem to restrain optimization procedures and render standardization techniques for enhanced practical applications complex. Hence, results of laboratory trials are not usually consistent with real-life applications. With the growing field of protein engineering, these complexities can be overcome by modifying or redesigning chitinases to enhance specific features required for specific applications. In this review, the variations in features and mechanisms of chitinases that limit their exploitation in biotechnological applications are compiled. Recent attempts to engineer chitinases for improved efficiency are also highlighted.
Keywords: Chitinases; biocontrol; chitooligosaccharides; protein engineering; variability.
© 2018 The Author(s).
Figures
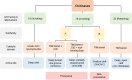
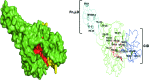


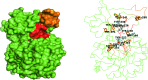
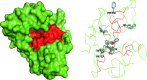


References
-
- Hamed I., Özogul F. and Regenstein J.M. (2016) Industrial applications of crustacean by-products (chitin, chitosan, and chitooligosaccharides): a review. Trends Food Sci. Technol. 48, 40–50 10.1016/j.tifs.2015.11.007 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

