A Large Open Pangenome and a Small Core Genome for Giant Pandoraviruses
- PMID: 30042742
- PMCID: PMC6048876
- DOI: 10.3389/fmicb.2018.01486
A Large Open Pangenome and a Small Core Genome for Giant Pandoraviruses
Abstract
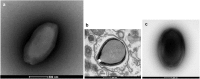
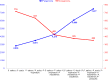
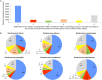
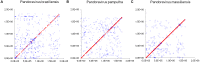
Giant viruses of amoebae are distinct from classical viruses by the giant size of their virions and genomes. Pandoraviruses are the record holders in size of genomes and number of predicted genes. Three strains, P. salinus, P. dulcis, and P. inopinatum, have been described to date. We isolated three new ones, namely P. massiliensis, P. braziliensis, and P. pampulha, from environmental samples collected in Brazil. We describe here their genomes, the transcriptome and proteome of P. massiliensis, and the pangenome of the group encompassing the six pandoravirus isolates. Genome sequencing was performed with an Illumina MiSeq instrument. Genome annotation was performed using GeneMarkS and Prodigal softwares and comparative genomic analyses. The core genome and pangenome were determined using notably ProteinOrtho and CD-HIT programs. Transcriptomics was performed for P. massiliensis with the Illumina MiSeq instrument; proteomics was also performed for this virus using 1D/2D gel electrophoresis and mass spectrometry on a Synapt G2Si Q-TOF traveling wave mobility spectrometer. The genomes of the three new pandoraviruses are comprised between 1.6 and 1.8 Mbp. The genomes of P. massiliensis, P. pampulha, and P. braziliensis were predicted to harbor 1,414, 2,368, and 2,696 genes, respectively. These genes comprise up to 67% of ORFans. Phylogenomic analyses showed that P. massiliensis and P. braziliensis were more closely related to each other than to the other pandoraviruses. The core genome of pandoraviruses comprises 352 clusters of genes, and the ratio core genome/pangenome is less than 0.05. The extinction curve shows clearly that the pangenome is still open. A quarter of the gene content of P. massiliensis was detected by transcriptomics. In addition, a product for a total of 162 open reading frames were found by proteomic analysis of P. massiliensis virions, including notably the products of 28 ORFans, 99 hypothetical proteins, and 90 core genes. Further analyses should allow to gain a better knowledge and understanding of the evolution and origin of these giant pandoraviruses, and of their relationships with viruses and cellular microorganisms.
Keywords: core genome; giant virus; megavirales; pandoravirus; pangenome.
Figures






References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

