Characterization of NDM-Encoding Plasmids From Enterobacteriaceae Recovered From Czech Hospitals
- PMID: 30042758
- PMCID: PMC6048247
- DOI: 10.3389/fmicb.2018.01549
Characterization of NDM-Encoding Plasmids From Enterobacteriaceae Recovered From Czech Hospitals
Abstract
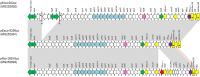
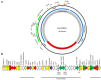
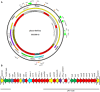
The aim of the present study was to characterize sporadic cases and an outbreak of NDM-like-producing Enterobacteriaceae recovered from hospital settings, in Czechia. During 2016, 18 Entrobacteriaceae isolates including 10 Enterobacter cloacae complex (9 E. xiangfangensis and 1 E. asburiae), 4 Escherichia coli, 1 Kluyvera intermedia, 1 Klebsiella pneumoniae, 1 Klebsiella oxytoca, and 1 Raoultella ornithinolytica that produced NDM-like carbapenemases were isolated from 15 patients. Three of the patients were colonized or infected by two different NDM-like producers. Moreover, an NDM-4-producing isolate of E. cloacae complex, isolated in 2012, was studied for comparative purposes. All isolates of E. cloacae complex, except the E. asburiae, recovered from the same hospital, were assigned to ST182. Additionally, two E. coli belonged to ST167, while the remaining isolates were not clonally related. Thirteen isolates carried blaNDM-4, while six isolates carried blaNDM-1 (n = 3) or blaNDM-5 (n = 3). Almost all isolates carried blaNDM-like-carrying plasmids being positive for the IncX3 allele, except ST58 E. coli and ST14 K. pneumoniae isolates producing NDM-1. Analysis of plasmid sequences revealed that all IncX3 blaNDM-like-carrying plasmids exhibited a high similarity to each other and to previously described plasmids, like pNDM-QD28, reported from worldwide. However, NDM-4-encoding plasmids differed from other IncX3 plasmids by the insertion of a Tn3-like transposon. On the other hand, the ST58 E. coli and ST14 K. pneumoniae isolates carried two novel NDM-1-encoding plasmids, pKpn-35963cz, and pEsco-36073cz. Plasmid pKpn-35963cz that was an IncFIB(K) molecule contained an acquired sequence, encoding NDM-1 metallo-β-lactamase (MβL), which exhibited high similarity to the mosaic region of pS-3002cz from an ST11 K. pneumoniae from Czechia. Finally, pEsco-36073cz was a multireplicon A/C2+R NDM-1-encoding plasmid. Similar to other type 1 A/C2 plasmids, the blaNDM-1 gene was located within the ARI-A resistance island. These findings underlined that IncX3 plasmids have played a major role in the dissemination of blaNDM-like genes in Czech hospitals. In combination with further evolvement of NDM-like-encoding MDR plasmids through reshuffling, NDM-like producers pose an important public threat.
Keywords: Enterobacter xiangfangensis; IncX3; NDM; ST182; metallo-β-lactamases.
Figures
References
-
- Bocanegra-Ibarias P., Garza-González E., Morfín-Otero R., Barrios H., Villarreal-Treviño L., Rodríguez-Noriega E., et al. . (2017). Molecular and microbiological report of a hospital outbreak of NDM-1-carrying Enterobacteriaceae in Mexico. PLoS ONE 12:e0179651. 10.1371/journal.pone.0179651 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

