Inosine Monophosphate Dehydrogenase Dependence in a Subset of Small Cell Lung Cancers
- PMID: 30043754
- PMCID: PMC6125205
- DOI: 10.1016/j.cmet.2018.06.005
Inosine Monophosphate Dehydrogenase Dependence in a Subset of Small Cell Lung Cancers
Abstract
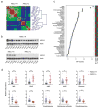
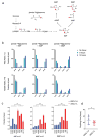
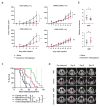
Small cell lung cancer (SCLC) is a rapidly lethal disease with few therapeutic options. We studied metabolic heterogeneity in SCLC to identify subtype-selective vulnerabilities. Metabolomics in SCLC cell lines identified two groups correlating with high or low expression of the Achaete-scute homolog-1 (ASCL1) transcription factor (ASCL1High and ASCL1Low), a lineage oncogene. Guanosine nucleotides were elevated in ASCL1Low cells and tumors from genetically engineered mice. ASCL1Low tumors abundantly express the guanosine biosynthetic enzymes inosine monophosphate dehydrogenase-1 and -2 (IMPDH1 and IMPDH2). These enzymes are transcriptional targets of MYC, which is selectively overexpressed in ASCL1Low SCLC. IMPDH inhibition reduced RNA polymerase I-dependent expression of pre-ribosomal RNA and potently suppressed ASCL1Low cell growth in culture, selectively reduced growth of ASCL1Low xenografts, and combined with chemotherapy to improve survival in genetic mouse models of ASCL1Low/MYCHigh SCLC. The data define an SCLC subtype-selective vulnerability related to dependence on de novo guanosine nucleotide synthesis.
Keywords: IMPDH; lung cancer; metabolism; metabolomics; therapeutics.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
R.J.D. is an advisor for Agios Pharmaceuticals.
Figures




References
-
- Akiyama T, Okazaki H, Takahashi K, Hasegawa A, Tanabe K, Uchida K, Takahara S, Toma H. Mizoribine in combination therapy with tacrolimus for living donor renal transplantation: analysis of a nationwide study in Japan. Transplant Proc. 2005;37:843–845. - PubMed
-
- Antonia SJ, Lopez-Martin JA, Bendell J, Ott PA, Taylor M, Eder JP, Jager D, Pietanza MC, Le DT, de Braud F, et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. The Lancet Oncology. 2016;17:883–895. - PubMed
-
- Augustyn A, Borromeo M, Wang T, Fujimoto J, Shao C, Dospoy PD, Lee V, Tan C, Sullivan JP, Larsen JE, et al. ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:14788–14793. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

