Genome sequence and genetic transformation of a widely distributed and cultivated poplar
- PMID: 30044051
- PMCID: PMC6335071
- DOI: 10.1111/pbi.12989
Genome sequence and genetic transformation of a widely distributed and cultivated poplar
Abstract
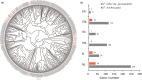
Populus alba is widely distributed and cultivated in Europe and Asia. This species has been used for diverse studies. In this study, we assembled a de novo genome sequence of P. alba var. pyramidalis (= P. bolleana) and confirmed its high transformation efficiency and short transformation time by experiments. Through a process of hybrid genome assembly, a total of 464 M of the genome was assembled. Annotation analyses predicted 37 901 protein-coding genes. This genome is highly collinear to that of P. trichocarpa, with most genes having orthologs in the two species. We found a marked expansion of gene families related to histone and the hormone auxin but loss of disease resistance genes in P. alba if compared with the closely related P. trichocarpa. The genome sequence presented here represents a valuable resource for further molecular functional analyses of this species as a new tree model, poplar breeding practices and comparative genomic analyses across different poplars.
Keywords: Populus alba; Populus bolleana; comparative genomics; gene families; transformation efficiency.
© 2018 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Boetzer, M. , Henkel, C.V. , Jansen, H.J. , Butler, D. and Pirovano, W. (2010) Scaffolding pre‐assembled contigs using SSPACE. Bioinformatics, 27, 578–579. - PubMed
-
- Bradshaw, H. , Ceulemans, R. , Davis, J. and Stettler, R. (2000) Emerging model systems in plant biology: poplar (Populus) as a model forest tree. J. Plant Growth Regul. 19, 306–313.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

