The HAUS Complex Is a Key Regulator of Non-centrosomal Microtubule Organization during Neuronal Development
- PMID: 30044976
- PMCID: PMC6083040
- DOI: 10.1016/j.celrep.2018.06.093
The HAUS Complex Is a Key Regulator of Non-centrosomal Microtubule Organization during Neuronal Development
Abstract
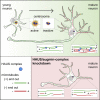
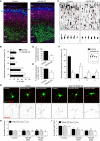
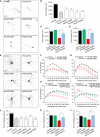
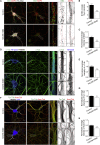
Neuron morphology and function are highly dependent on proper organization of the cytoskeleton. In neurons, the centrosome is inactivated early in development, and acentrosomal microtubules are generated by mechanisms that are poorly understood. Here, we show that neuronal migration, development, and polarization depend on the multi-subunit protein HAUS/augmin complex, previously described to be required for mitotic spindle assembly in dividing cells. The HAUS complex is essential for neuronal microtubule organization by ensuring uniform microtubule polarity in axons and regulation of microtubule density in dendrites. Using live-cell imaging and high-resolution microscopy, we found that distinct HAUS clusters are distributed throughout neurons and colocalize with γ-TuRC, suggesting local microtubule nucleation events. We propose that the HAUS complex locally regulates microtubule nucleation events to control proper neuronal development.
Keywords: actin; centrosome; development; microtubule; migration; neuron; nucleation.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
References
-
- Akhmanova A., Hoogenraad C.C. Microtubule minus-end-targeting proteins. Curr. Biol. 2015;25:R162–R171. - PubMed
-
- Baas P.W., Karabay A., Qiang L. Microtubules cut and run. Trends Cell Biol. 2005;15:518–524. - PubMed
-
- Conde C., Cáceres A. Microtubule assembly, organization and dynamics in axons and dendrites. Nat. Rev. Neurosci. 2009;10:319–332. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

