Human fibroblast and stem cell resource from the Dominantly Inherited Alzheimer Network
- PMID: 30045758
- PMCID: PMC6060509
- DOI: 10.1186/s13195-018-0400-0
Human fibroblast and stem cell resource from the Dominantly Inherited Alzheimer Network
Abstract
Background: Mutations in amyloid precursor protein (APP), presenilin 1 (PSEN1) and presenilin 2 (PSEN2) cause autosomal dominant forms of Alzheimer disease (ADAD). More than 280 pathogenic mutations have been reported in APP, PSEN1, and PSEN2. However, understanding of the basic biological mechanisms that drive the disease are limited. The Dominantly Inherited Alzheimer Network (DIAN) is an international observational study of APP, PSEN1, and PSEN2 mutation carriers with the goal of determining the sequence of changes in presymptomatic mutation carriers who are destined to develop Alzheimer disease.
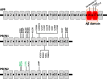
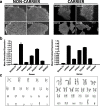
Results: We generated a library of 98 dermal fibroblast lines from 42 ADAD families enrolled in DIAN. We have reprogrammed a subset of the DIAN fibroblast lines into patient-specific induced pluripotent stem cell (iPSC) lines. These cells were thoroughly characterized for pluripotency markers.
Conclusions: This library represents a comprehensive resource that can be used for disease modeling and the development of novel therapeutics.
Keywords: Amyloid precursor protein; Dominantly Inherited Alzheimer Network; Fibroblasts; Induced pluripotent stem cells; Presenilin 1; Presenilin 2.
Conflict of interest statement
Ethics approval and consent to participate
The Washington University IRB reviewed the study protocol (IRB no. 201104178, 201306108). All subjects included in this study, or their proxies, gave written informed consent.
Consent for publication
Not applicable.
Competing interests
AMG is a member of the scientific advisory board for Denali Therapeutics and serves on the Genetic Scientific Advisory Panel for Pfizer. RJB receives laboratory research funding from the National Institutes of Health, Alzheimer’s Association, BrightFocus Foundation, Rainwater Foundation Tau Consortium, Association for Frontotemporal Degeneration, the Cure Alzheimer’s Fund, and the Tau SILK Consortium (AbbVie, Biogen, and Eli Lilly and Co.). Funding for clinical trials not related to this research include the National Institutes of Health, Alzheimer’s Association, Eli Lilly and Co., Hoffman-La Roche, Janssen, Avid Radiopharmaceuticals, GHR Foundation, and an anonymous foundation. RJB also receives research funding from the DIAN Pharma Consortium (AbbVie, Amgen, AstraZeneca, Biogen, Eisai, Eli Lilly and Co., Hoffman-La Roche, Janssen, Pfizer, and Sanofi). RJB has received honoraria from Janssen and Pfizer as a speaker and from Merck and Pfizer as an advisory board member. Washington University, RJB, and DH have equity ownership interests in C2N Diagnostics and receive royalty income based on technology (stable isotope labeling kinetics and blood plasma assay) licensed by Washington University to C2N Diagnostics. RJB receives income from C2N Diagnostics for serving on its scientific advisory board. Washington University, with RJB as coinventor, has submitted the U.S. nonprovisional patent application “Methods for Measuring the Metabolism of CNS Derived Biomolecules In Vivo” and a provisional patent application, “Plasma Based Methods for Detecting CNS Amyloid Deposition.” The remaining authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

