Unperturbed expression bias of imprinted genes in schizophrenia
- PMID: 30046039
- PMCID: PMC6060121
- DOI: 10.1038/s41467-018-04960-9
Unperturbed expression bias of imprinted genes in schizophrenia
Abstract
How gene expression correlates with schizophrenia across individuals is beginning to be examined through analyses of RNA-seq from postmortem brains of individuals with disease and control brains. Here we focus on variation in allele-specific expression, following up on the CommonMind Consortium (CMC) RNA-seq experiments of nearly 600 human dorsolateral prefrontal cortex (DLPFC) samples. Analyzing the extent of allelic expression bias-a hallmark of imprinting-we find that the number of imprinted human genes is consistent with lower estimates (≈0.5% of all genes), and thus contradicts much higher estimates. Moreover, the handful of putatively imprinted genes are all in close genomic proximity to known imprinted genes. Joint analysis of the imprinted genes across hundreds of individuals allowed us to establish how allelic bias depends on various factors. We find that age and genetic ancestry have gene-specific, differential effect on allelic bias. In contrast, allelic bias appears to be independent of schizophrenia.
Conflict of interest statement
The authors declare no competing interests.
Figures

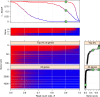

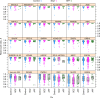

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

