Differential regulation of the Wnt/β-catenin pathway by hepatitis C virus recombinants expressing core from various genotypes
- PMID: 30046100
- PMCID: PMC6060129
- DOI: 10.1038/s41598-018-29078-2
Differential regulation of the Wnt/β-catenin pathway by hepatitis C virus recombinants expressing core from various genotypes
Abstract
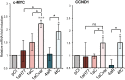
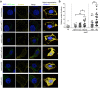
Clinical studies have suggested association of some hepatitis C virus (HCV) subtypes or isolates with progression toward hepatocellular carcinoma (HCC). HCV core protein has been reported to interfere with host Wnt/β-catenin pathway, a cell fate-determining pathway, which plays a major role in HCC. Here, we investigated the impact of HCV core genetic variability in the dysregulation of Wnt/β-catenin pathway. We used both transient expression of core proteins from clinical isolates of HCV subtypes 1a (Cambodia), 4a (Romania) and 4f (Cameroon) and infection systems based on a set of engineered intergenotypic recombinant viruses encoding core from these various clinical strains. We found that TCF transcription factor-dependent reporter activity was upregulated by core in a strain-specific manner. We documented core sequence-specific transcriptional upregulation of several β-catenin downstream target genes associated with cell proliferation and malignant transformation, fibrogenesis or fat accumulation. The extent of β-catenin nuclear translocation varied in accordance with β-catenin downstream gene upregulation in infected cells. Pairwise comparisons of subgenotypic core recombinants and mutated core variants unveiled the critical role of core residues 64 and 71 in these dysregulations. In conclusion, this work identified natural core polymorphisms involved in HCV strain-specific activation of Wnt/β-catenin pathway in relevant infection systems.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Moradpour D, Penin F. Hepatitis C virus proteins: from structure to function. Curr Top Microbiol Immunol. 2013;369:113–142. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

