53BP1 cooperation with the REV7-shieldin complex underpins DNA structure-specific NHEJ
- PMID: 30046110
- PMCID: PMC6989217
- DOI: 10.1038/s41586-018-0362-1
53BP1 cooperation with the REV7-shieldin complex underpins DNA structure-specific NHEJ
Abstract
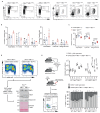
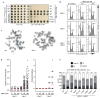
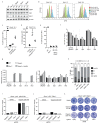
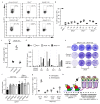
53BP1 governs a specialized, context-specific branch of the classical non-homologous end joining DNA double-strand break repair pathway. Mice lacking 53bp1 (also known as Trp53bp1) are immunodeficient owing to a complete loss of immunoglobulin class-switch recombination1,2, and reduced fidelity of long-range V(D)J recombination3. The 53BP1-dependent pathway is also responsible for pathological joining events at dysfunctional telomeres4, and its unrestricted activity in Brca1-deficient cellular and tumour models causes genomic instability and oncogenesis5-7. Cells that lack core non-homologous end joining proteins are profoundly radiosensitive8, unlike 53BP1-deficient cells9,10, which suggests that 53BP1 and its co-factors act on specific DNA substrates. Here we show that 53BP1 cooperates with its downstream effector protein REV7 to promote non-homologous end joining during class-switch recombination, but REV7 is not required for 53BP1-dependent V(D)J recombination. We identify shieldin-a four-subunit putative single-stranded DNA-binding complex comprising REV7, c20orf196 (SHLD1), FAM35A (SHLD2) and FLJ26957 (SHLD3)-as the factor that explains this specificity. Shieldin is essential for REV7-dependent DNA end-protection and non-homologous end joining during class-switch recombination, and supports toxic non-homologous end joining in Brca1-deficient cells, yet is dispensable for REV7-dependent interstrand cross-link repair. The 53BP1 pathway therefore comprises distinct double-strand break repair activities within chromatin and single-stranded DNA compartments, which explains both the immunological differences between 53bp1- and Rev7- deficient mice and the context specificity of the pathway.
Figures






References
-
- Manis JP, et al. 53BP1 links DNA damage-response pathways to immunoglobulin heavy chain class-switch recombination. Nat Immunol. 2004;5:481–487. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

