Identification of integrin drug targets for 17 solid tumor types
- PMID: 30046394
- PMCID: PMC6059022
- DOI: 10.18632/oncotarget.25731
Identification of integrin drug targets for 17 solid tumor types
Abstract
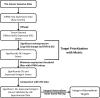
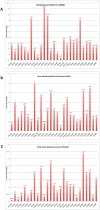
Integrins are contributors to remodeling of the extracellular matrix and cell migration. Integrins participate in the assembly of the actin cytoskeleton, regulate growth factor signaling pathways, cell proliferation, and control cell motility. In solid tumors, integrins are involved in promoting metastasis to distant sites, and angiogenesis. Integrins are a key target in cancer therapy and imaging. Integrin antagonists have proven successful in halting invasion and migration of tumors. Overexpressed integrins are prime anti-cancer drug targets. To streamline the development of specific integrin cancer therapeutics, we curated data to predict which integrin heterodimers are pausible therapeutic targets against 17 different solid tumors. Computational analysis of The Cancer Genome Atlas (TCGA) gene expression data revealed a set of integrin targets that are differentially expressed in tumors. Filtered by FPKM (Fragments Per Kilobase of transcript per Million mapped reads) expression level, overexpressed subunits were paired into heterodimeric protein targets. By comparing the RNA-seq differential expression results with immunohistochemistry (IHC) data, overexpressed integrin subunits were validated. Biologics and small molecule drug compounds against these identified overexpressed subunits and heterodimeric receptors are potential therapeutics against these cancers. In addition, high-affinity and high-specificity ligands against these integrins can serve as efficient vehicles for delivery of cancer drugs, nanotherapeutics, or imaging probes against cancer.
Keywords: computational genomics; integrins; precision medicine; therapeutic target selection; transcriptomics.
Conflict of interest statement
CONFLICTS OF INTEREST All authors declare that there are no conflicts of interest.
Figures






References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

