Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies
- PMID: 30048243
- PMCID: PMC6075434
- DOI: 10.18632/aging.101508
Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies
Abstract
DNA methylation (DNAm)-based biomarkers of aging have been developed for many tissues and organs. However, these biomarkers have sub-optimal accuracy in fibroblasts and other cell types used in ex vivo studies. To address this challenge, we developed a novel and highly robust DNAm age estimator (based on 391 CpGs) for human fibroblasts, keratinocytes, buccal cells, endothelial cells, lymphoblastoid cells, skin, blood, and saliva samples. High age correlations can also be observed in sorted neurons, glia, brain, liver, and even bone samples. Gestational age correlates with DNAm age in cord blood. When used on fibroblasts from Hutchinson Gilford Progeria Syndrome patients, this age estimator (referred to as the skin & blood clock) uncovered an epigenetic age acceleration with a magnitude that is below the sensitivity levels of other DNAm-based biomarkers. Furthermore, this highly sensitive age estimator accurately tracked the dynamic aging of cells cultured ex vivo and revealed that their proliferation is accompanied by a steady increase in epigenetic age. The skin & blood clock predicts lifespan and it relates to many age-related conditions. Overall, this biomarker is expected to become useful for forensic applications (e.g. blood or buccal swabs) and for a quantitative ex vivo human cell aging assay.
Keywords: DNA methylation; Hutchinson-Gilford; epigenetics; fibroblasts; progeria.
Conflict of interest statement
Figures

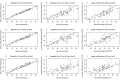

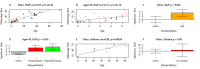
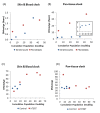
Comment in
-
Epigenetic clocks galore: a new improved clock predicts age-acceleration in Hutchinson Gilford Progeria Syndrome patients.Aging (Albany NY). 2018 Aug 21;10(8):1799-1800. doi: 10.18632/aging.101533. Aging (Albany NY). 2018. PMID: 30130743 Free PMC article. No abstract available.
-
The epigenetic clock: a molecular crystal ball for human aging?Aging (Albany NY). 2019 Jan 21;11(2):833-835. doi: 10.18632/aging.101712. Aging (Albany NY). 2019. PMID: 30669120 Free PMC article. No abstract available.
References
-
- Rakyan VK, Down TA, Maslau S, Andrew T, Yang TP, Beyan H, Whittaker P, McCann OT, Finer S, Valdes AM, Leslie RD, Deloukas P, Spector TD. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res. 2010; 20:434–39. 10.1101/gr.103101.109 - DOI - PMC - PubMed
-
- Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, Campan M, Noushmehr H, Bell CG, Maxwell AP, Savage DA, Mueller-Holzner E, Marth C, et al.. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010; 20:440–46. 10.1101/gr.103606.109 - DOI - PMC - PubMed
MeSH terms
Grants and funding
- HHSN268201600018C/HL/NHLBI NIH HHS/United States
- HHSN268201300048C/HL/NHLBI NIH HHS/United States
- R01 CA210916/CA/NCI NIH HHS/United States
- HHSN268201300049C/HL/NHLBI NIH HHS/United States
- HHSN268201600003C/HL/NHLBI NIH HHS/United States
- HHSN268201600001C/HL/NHLBI NIH HHS/United States
- HHSN268201300046C/HL/NHLBI NIH HHS/United States
- U34 AG051425/AG/NIA NIH HHS/United States
- U54 GM115428/GM/NIGMS NIH HHS/United States
- U01 AG060908/AG/NIA NIH HHS/United States
- HHSN268201600002C/HL/NHLBI NIH HHS/United States
- R01 HL116446/HL/NHLBI NIH HHS/United States
- R01 ES020836/ES/NIEHS NIH HHS/United States
- HHSN268201300047C/HL/NHLBI NIH HHS/United States
- HHSN268201300050C/HL/NHLBI NIH HHS/United States
- HHSN268201600004C/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

