Identification of 613 new loci associated with heel bone mineral density and a polygenic risk score for bone mineral density, osteoporosis and fracture
- PMID: 30048462
- PMCID: PMC6062019
- DOI: 10.1371/journal.pone.0200785
Identification of 613 new loci associated with heel bone mineral density and a polygenic risk score for bone mineral density, osteoporosis and fracture
Erratum in
-
Correction: Identification of 613 new loci associated with heel bone mineral density and a polygenic risk score for bone mineral density, osteoporosis and fracture.PLoS One. 2019 Mar 13;14(3):e0213962. doi: 10.1371/journal.pone.0213962. eCollection 2019. PLoS One. 2019. PMID: 30865700 Free PMC article.
Abstract
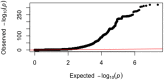
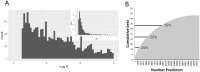
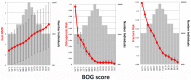
Low bone mineral density (BMD) leads to osteoporosis, and is a risk factor for bone fractures, including stress fractures. Using data from UK Biobank, a genome-wide association study identified 1,362 independent SNPs that clustered into 899 loci of which 613 are new. These data were used to train a genetic algorithm using 22,886 SNPs as predictors and showing a correlation with heel bone mineral density of 0.415. Combining this genetic algorithm with height, weight, age and sex resulted in a correlation with heel bone mineral density of 0.496. Individuals with low scores (2.2% of total) showed a change in BMD of -1.16 T-score units, an increase in risk for osteoporosis of 17.4 fold and an increase in risk for fracture of 1.87 fold. Genetic predictors could assist in the identification of individuals at risk for osteoporosis or fractures.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
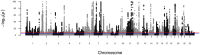



Similar articles
-
Genetic determinants of heel bone properties: genome-wide association meta-analysis and replication in the GEFOS/GENOMOS consortium.Hum Mol Genet. 2014 Jun 1;23(11):3054-68. doi: 10.1093/hmg/ddt675. Epub 2014 Jan 14. Hum Mol Genet. 2014. PMID: 24430505 Free PMC article.
-
Hyperthyroidism and bone mineral density: Dissecting the causal association with Mendelian randomization analysis.Clin Endocrinol (Oxf). 2021 Jan;94(1):119-127. doi: 10.1111/cen.14330. Epub 2020 Sep 28. Clin Endocrinol (Oxf). 2021. PMID: 32947644
-
Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study.Lancet. 2008 May 3;371(9623):1505-12. doi: 10.1016/S0140-6736(08)60599-1. Lancet. 2008. PMID: 18455228 Free PMC article.
-
Dissecting the Genetics of Osteoporosis using Systems Approaches.Trends Genet. 2019 Jan;35(1):55-67. doi: 10.1016/j.tig.2018.10.004. Epub 2018 Nov 20. Trends Genet. 2019. PMID: 30470485 Free PMC article. Review.
-
Collaborative meta-analysis: associations of 150 candidate genes with osteoporosis and osteoporotic fracture.Ann Intern Med. 2009 Oct 20;151(8):528-37. doi: 10.7326/0003-4819-151-8-200910200-00006. Ann Intern Med. 2009. PMID: 19841454 Free PMC article.
Cited by
-
Influence of glycoprotein MUC1 on trafficking of the Ca2+-selective ion channels, TRPV5 and TRPV6, and on in vivo calcium homeostasis.J Biol Chem. 2023 Mar;299(3):102925. doi: 10.1016/j.jbc.2023.102925. Epub 2023 Jan 20. J Biol Chem. 2023. PMID: 36682497 Free PMC article.
-
Thousands of scientists publish a paper every five days.Nature. 2018 Sep;561(7722):167-169. doi: 10.1038/d41586-018-06185-8. Nature. 2018. PMID: 30209384 No abstract available.
-
Estrogen receptor alpha and NFATc1 bind to a bone mineral density-associated SNP to repress WNT5B in osteoblasts.Am J Hum Genet. 2022 Jan 6;109(1):97-115. doi: 10.1016/j.ajhg.2021.11.018. Epub 2021 Dec 13. Am J Hum Genet. 2022. PMID: 34906330 Free PMC article.
-
Cis-meQTL for cocaine use-associated DNA methylation in an HIV-positive cohort show pleiotropic effects on multiple traits.BMC Genomics. 2023 Sep 20;24(1):556. doi: 10.1186/s12864-023-09661-2. BMC Genomics. 2023. PMID: 37730558 Free PMC article.
-
Analysis of independent cohorts of outbred CFW mice reveals novel loci for behavioral and physiological traits and identifies factors determining reproducibility.G3 (Bethesda). 2022 Jan 4;12(1):jkab394. doi: 10.1093/g3journal/jkab394. G3 (Bethesda). 2022. PMID: 34791208 Free PMC article.
References
-
- Gluer CC, Eastell R, Reid DM, Felsenberg D, Roux C, Barkmann R, et al. (2004) Association of five quantitative ultrasound devices and bone densitometry with osteoporotic vertebral fractures in a population-based sample: the OPUS Study. J Bone Miner Res 19: 782–793. 10.1359/JBMR.040304 - DOI - PubMed
-
- Nattiv A (2000) Stress fractures and bone health in track and field athletes. J Sci Med Sport 3: 268–279. - PubMed
-
- Jones BH, Knapik JJ (1999) Physical training and exercise-related injuries. Surveillance, research and injury prevention in military populations. Sports Med 27: 111–125. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

