Automatic extraction of gene-disease associations from literature using joint ensemble learning
- PMID: 30048465
- PMCID: PMC6061985
- DOI: 10.1371/journal.pone.0200699
Automatic extraction of gene-disease associations from literature using joint ensemble learning
Abstract
A wealth of knowledge concerning relations between genes and its associated diseases is present in biomedical literature. Mining these biological associations from literature can provide immense support to research ranging from drug-targetable pathways to biomarker discovery. However, time and cost of manual curation heavily slows it down. In this current scenario one of the crucial technologies is biomedical text mining, and relation extraction shows the promising result to explore the research of genes associated with diseases. By developing automatic extraction of gene-disease associations from the literature using joint ensemble learning we addressed this problem from a text mining perspective. In the proposed work, we employ a supervised machine learning approach in which a rich feature set covering conceptual, syntax and semantic properties jointly learned with word embedding are trained using ensemble support vector machine for extracting gene-disease relations from four gold standard corpora. Upon evaluating the machine learning approach shows promised results of 85.34%, 83.93%,87.39% and 85.57% of F-measure on EUADR, GAD, CoMAGC and PolySearch corpora respectively. We strongly believe that the presented novel approach combining rich syntax and semantic feature set with domain-specific word embedding through ensemble support vector machines evaluated on four gold standard corpora can act as a new baseline for future works in gene-disease relation extraction from literature.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
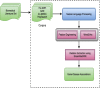
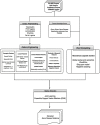

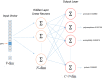
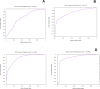
References
-
- Ware M, Mabe M. The STM report: An overview of scientific and scholarly journal publishing. 2015.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

