Antigenic and genetic characterization of influenza viruses isolated in Mozambique during the 2015 season
- PMID: 30048502
- PMCID: PMC6062064
- DOI: 10.1371/journal.pone.0201248
Antigenic and genetic characterization of influenza viruses isolated in Mozambique during the 2015 season
Abstract
Background: Due to the high rate of antigenic variation of influenza virus, seasonal characterization of the virus is crucial to assess and monitor the emergence of new pathogenic variants and hence formulate effective control measures. However, no study has yet been conducted in Mozambique to assess genetic, antigenic and antiviral susceptibility profile of influenza virus.
Methods: A subset of samples (n = 20) from influenza positive children detected in two hospitals in Maputo city during 2015 season as part of the implementation of influenza surveillance system, were selected. The following assays were performed on these samples: antigenic characterization by hemagglutination inhibition assay, genetic characterization by Sanger sequencing of hemagglutinin (HA) and neuraminidase (NA) and susceptibility to oseltamivir and zanamivir (NA inhibitors) by enzymatic assay.
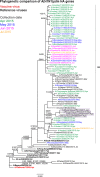
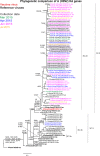
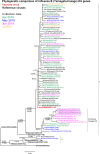
Results: The A(H1N1)pdm09 subtype viruses remained closely related antigenically and genetically to the 2016 vaccine virus A/California/7/2009 and other widely distributed viruses belonging to genetic group 6B. The majority of influenza A(H3N2) viruses studied were antigenically similar to the 2016-2017 vaccine virus, A/Hong Kong/4801/2014, and their HA and NA gene sequences fell into genetic subclade 3C.2a being closely related to viruses circulating in southern Africa. The influenza B viruses were antigenically similar to the 2016 season vaccine virus and HA sequences of all three fell into the B/Yamagata-lineage, clade 3, but contained NA genes of the B/Victoria-lineage. All tested viruses were sensitive to oseltamivir and zanamivir.
Conclusion: Overall, all Mozambican influenza A and B viruses were most closely related to Southern African viruses and all were sensitive to oseltamivir and zanamivir. These findings suggest the existence of an ecological niche of influenza viruses within the region and hence highlighting the need for joint epidemiologic and virologic surveillance to monitor the evolution of influenza viruses.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- World Health Organization. Influenza (Seasonal): fact sheet . In: World Health Organization; [Internet]. 2014. pp. 1–4.
-
- Shaw ML, Palese P. Orthomyxoviridae. Fields Virology. 2013. pp. 1151–1185. doi:9781451105636
-
- Payne S. Family Orthomyxoviridae. Viruses. 2017. pp. 197–208. 10.1016/B978-0-12-803109-4.00023–4 - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

