Mutant Muscle LIM Protein C58G causes cardiomyopathy through protein depletion
- PMID: 30048712
- PMCID: PMC6117453
- DOI: 10.1016/j.yjmcc.2018.07.248
Mutant Muscle LIM Protein C58G causes cardiomyopathy through protein depletion
Abstract
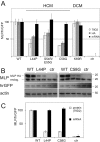
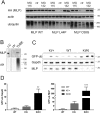
Cysteine and glycine rich protein 3 (CSRP3) encodes Muscle LIM Protein (MLP), a well-established disease gene for Hypertrophic Cardiomyopathy (HCM). MLP, in contrast to the proteins encoded by the other recognised HCM disease genes, is non-sarcomeric, and has important signalling functions in cardiomyocytes. To gain insight into the disease mechanisms involved, we generated a knock-in mouse (KI) model, carrying the well documented HCM-causing CSRP3 mutation C58G. In vivo phenotyping of homozygous KI/KI mice revealed a robust cardiomyopathy phenotype with diastolic and systolic left ventricular dysfunction, which was supported by increased heart weight measurements. Transcriptome analysis by RNA-seq identified activation of pro-fibrotic signalling, induction of the fetal gene programme and activation of markers of hypertrophic signalling in these hearts. Further ex vivo analyses validated the activation of these pathways at transcript and protein level. Intriguingly, the abundance of MLP decreased in KI/KI mice by 80% and in KI/+ mice by 50%. Protein depletion was also observed in cellular studies for two further HCM-causing CSRP3 mutations (L44P and S54R/E55G). We show that MLP depletion is caused by proteasome action. Moreover, MLP C58G interacts with Bag3 and results in a proteotoxic response in the homozygous knock-in mice, as shown by induction of Bag3 and associated heat shock proteins. In conclusion, the newly generated mouse model provides insights into the underlying disease mechanisms of cardiomyopathy caused by mutations in the non-sarcomeric protein MLP. Furthermore, our cellular experiments suggest that protein depletion and proteasomal overload also play a role in other HCM-causing CSPR3 mutations that we investigated, indicating that reduced levels of functional MLP may be a common mechanism for HCM-causing CSPR3 mutations.
Keywords: Hypertrophic cardiomyopathy; In vivo phenotyping; MLP C58G mutation; Mouse knock-in model; Proteasome; Protein depletion; RNAseq transcriptome analysis; Sarcomere.
Copyright © 2018 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures





References
-
- Arber S., Halder G., Caroni P. Muscle LIM protein, a novel essential regulator of myogenesis, promotes myogenic differentiation. Cell. 1994;79(2):221–231. - PubMed
-
- Knoll R., Hoshijima M., Hoffman H.M., Person V., Lorenzen-Schmidt I., Bang M.L., Hayashi T., Shiga N., Yasukawa H., Schaper W., McKenna W., Yokoyama M., Schork N.J., Omens J.H., McCulloch A.D., Kimura A., Gregorio C.C., Poller W., Schaper J., Schultheiss H.P., Chien K.R. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111(7):943–955. - PubMed
-
- Geier C., Gehmlich K., Ehler E., Hassfeld S., Perrot A., Hayess K., Cardim N., Wenzel K., Erdmann B., Krackhardt F., Posch M.G., Osterziel K.J., Bublak A., Nagele H., Scheffold T., Dietz R., Chien K.R., Spuler S., Furst D.O., Nurnberg P., Ozcelik C. Beyond the sarcomere: CSRP3 mutations cause hypertrophic cardiomyopathy. Hum. Mol. Genet. 2008;17(18):2753–2765. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BB/P001599/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- RE/13/1/30181/BHF_/British Heart Foundation/United Kingdom
- BB/R001049/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/M007103/1 /BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 201543/B/16/Z/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

