Patient-derived Models of Abiraterone- and Enzalutamide-resistant Prostate Cancer Reveal Sensitivity to Ribosome-directed Therapy
- PMID: 30049486
- PMCID: PMC6351078
- DOI: 10.1016/j.eururo.2018.06.020
Patient-derived Models of Abiraterone- and Enzalutamide-resistant Prostate Cancer Reveal Sensitivity to Ribosome-directed Therapy
Abstract
Background: The intractability of castration-resistant prostate cancer (CRPC) is exacerbated by tumour heterogeneity, including diverse alterations to the androgen receptor (AR) axis and AR-independent phenotypes. The availability of additional models encompassing this heterogeneity would facilitate the identification of more effective therapies for CRPC.
Objective: To discover therapeutic strategies by exploiting patient-derived models that exemplify the heterogeneity of CRPC.
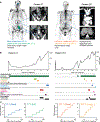
Design, setting, and participants: Four new patient-derived xenografts (PDXs) were established from independent metastases of two patients and characterised using integrative genomics. A panel of rationally selected drugs was tested using an innovative ex vivo PDX culture system.
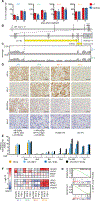
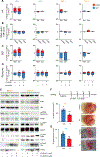
Intervention: The following drugs were evaluated: AR signalling inhibitors (enzalutamide and galeterone), a PARP inhibitor (talazoparib), a chemotherapeutic (cisplatin), a CDK4/6 inhibitor (ribociclib), bromodomain and extraterminal (BET) protein inhibitors (iBET151 and JQ1), and inhibitors of ribosome biogenesis/function (RNA polymerase I inhibitor CX-5461 and pan-PIM kinase inhibitor CX-6258).
Outcome measurements and statistical analysis: Drug efficacy in ex vivo cultures of PDX tissues was evaluated using immunohistochemistry for Ki67 and cleaved caspase-3 levels. Candidate drugs were also tested for antitumour efficacy in vivo, with tumour volume being the primary endpoint. Two-tailed t tests were used to compare drug and control treatments.
Results and limitations: Integrative genomics revealed that the new PDXs exhibited heterogeneous mechanisms of resistance, including known and novel AR mutations, genomic structural rearrangements of the AR gene, and a neuroendocrine-like AR-null phenotype. Despite their heterogeneity, all models were sensitive to the combination of ribosome-targeting agents CX-5461 and CX-6258.
Conclusions: This study demonstrates that ribosome-targeting drugs may be effective against diverse CRPC subtypes including AR-null disease, and highlights the potential of contemporary patient-derived models to prioritise treatment strategies for clinical translation.
Patient summary: Diverse types of therapy-resistant prostate cancers are sensitive to a new combination of drugs that inhibit protein synthesis pathways in cancer cells.
Keywords: Abiraterone; Androgen receptor; Castration-resistant prostate cancer; Enzalutamide; Explant; Neuroendocrine prostate cancer; Organoid; Patient-derived xenograft; Prostate cancer; Ribosome.
Copyright © 2018 European Association of Urology. Published by Elsevier B.V. All rights reserved.
Figures
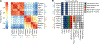

Comment in
-
Therapeutic Discovery for Castration Resistant Prostate Cancer: Patient-derived Xenograft Modelling Brings New Options to the Table.Eur Urol. 2018 Nov;74(5):573-574. doi: 10.1016/j.eururo.2018.06.049. Epub 2018 Jul 13. Eur Urol. 2018. PMID: 30017402 No abstract available.
References
-
- Sridhar SS, Freedland SJ, Gleave ME, et al. Castration-resistant prostate cancer: from new pathophysiology to new treatment. Eur Urol 2014;65:289–99. - PubMed
-
- Coutinho I, Day TK, Tilley WD, Selth LA. Androgen receptor signaling in castration-resistant prostate cancer: a lesson in persistence. Endocr Relat Cancer 2016;23:T179–97. - PubMed
-
- Li Y, Donmez N, Sahinalp C, et al. SRRM4 drives neuroendocrine transdifferentiation of prostate adenocarcinoma under androgen receptor pathway inhibition. Eur Urol 2017;71:68–78. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

