Rapid Paediatric Sequencing (RaPS): comprehensive real-life workflow for rapid diagnosis of critically ill children
- PMID: 30049826
- PMCID: PMC6252361
- DOI: 10.1136/jmedgenet-2018-105396
Rapid Paediatric Sequencing (RaPS): comprehensive real-life workflow for rapid diagnosis of critically ill children
Abstract
Background: Rare genetic conditions are frequent risk factors for, or direct causes of, paediatric intensive care unit (PICU) admission. Such conditions are frequently suspected but unidentified at PICU admission. Compassionate and effective care is greatly assisted by definitive diagnostic information. There is therefore a need to provide a rapid genetic diagnosis to inform clinical management.To date, whole genome sequencing (WGS) approaches have proved successful in diagnosing a proportion of children with rare diseases, but results may take months to report. Our aim was to develop an end-to-end workflow for the use of rapid WGS for diagnosis in critically ill children in a UK National Health Service (NHS) diagnostic setting.
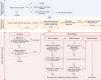
Methods: We sought to establish a multidisciplinary Rapid Paediatric Sequencing team for case selection, trio WGS, rapid bioinformatics sequence analysis and a phased analysis and reporting system to prioritise genes with a high likelihood of being causal.
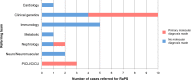
Results: Trio WGS in 24 critically ill children led to a molecular diagnosis in 10 (42%) through the identification of causative genetic variants. In 3 of these 10 individuals (30%), the diagnostic result had an immediate impact on the individual's clinical management. For the last 14 trios, the shortest time taken to reach a provisional diagnosis was 4 days (median 8.5 days).
Conclusion: Rapid WGS can be used to diagnose and inform management of critically ill children within the constraints of an NHS clinical diagnostic setting. We provide a robust workflow that will inform and facilitate the rollout of rapid genome sequencing in the NHS and other healthcare systems globally.
Keywords: genomics; paediatric intensive care unit; rapid diagnosis; rare disease; whole genome sequencing.
© Author(s) (or their employer(s)) 2018. Re-use permitted under CC BY. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures

References
-
- Borghesi A, Mencarelli MA, Memo L, Ferrero GB, Bartuli A, Genuardi M, Stronati M, Villani A, Renieri A, Corsello G. their respective Scientific Societies. Intersociety policy statement on the use of whole-exome sequencing in the critically ill newborn infant. Ital J Pediatr 2017;43:100 10.1186/s13052-017-0418-0 - DOI - PMC - PubMed
-
- Sawyer SL, Hartley T, Dyment DA, Beaulieu CL, Schwartzentruber J, Smith A, Bedford HM, Bernard G, Bernier FP, Brais B, Bulman DE, Warman Chardon J, Chitayat D, Deladoëy J, Fernandez BA, Frosk P, Geraghty MT, Gerull B, Gibson W, Gow RM, Graham GE, Green JS, Heon E, Horvath G, Innes AM, Jabado N, Kim RH, Koenekoop RK, Khan A, Lehmann OJ, Mendoza-Londono R, Michaud JL, Nikkel SM, Penney LS, Polychronakos C, Richer J, Rouleau GA, Samuels ME, Siu VM, Suchowersky O, Tarnopolsky MA, Yoon G, Zahir FR, Majewski J, Boycott KM. FORGE Canada Consortium Care4Rare Canada Consortium. Utility of whole-exome sequencing for those near the end of the diagnostic odyssey: time to address gaps in care. Clin Genet 2016;89:275–84. 10.1111/cge.12654 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
