Blueprints for Biosensors: Design, Limitations, and Applications
- PMID: 30050028
- PMCID: PMC6115959
- DOI: 10.3390/genes9080375
Blueprints for Biosensors: Design, Limitations, and Applications
Abstract
Biosensors are enabling major advances in the field of analytics that are both facilitating and being facilitated by advances in synthetic biology. The ability of biosensors to rapidly and specifically detect a wide range of molecules makes them highly relevant to a range of industrial, medical, ecological, and scientific applications. Approaches to biosensor design are as diverse as their applications, with major biosensor classes including nucleic acids, proteins, and transcription factors. Each of these biosensor types has advantages and limitations based on the intended application, and the parameters that are required for optimal performance. Specifically, the choice of biosensor design must consider factors such as the ligand specificity, sensitivity, dynamic range, functional range, mode of output, time of activation, ease of use, and ease of engineering. This review discusses the rationale for designing the major classes of biosensor in the context of their limitations and assesses their suitability to different areas of biotechnological application.
Keywords: analytics; aptamers; biosensors; high-throughput screening; metabolic engineering; molecular diagnostics; protein switches; synthetic biology.
Conflict of interest statement
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
Figures



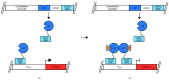

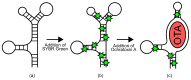
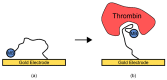

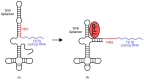
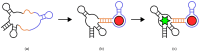


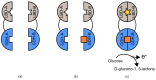

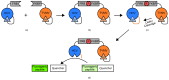
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

