Analysis of ex vivo HIV-1 infection in a controller-discordant couple
- PMID: 30050679
- PMCID: PMC6038135
- DOI: 10.1016/S2055-6640(20)30268-5
Analysis of ex vivo HIV-1 infection in a controller-discordant couple
Abstract
Objectives: Elite controllers (EC) are a rare group of individuals living with HIV-1 who naturally control HIV-1 replication to levels below the limit of detection without antiretroviral therapy (ART) and rarely progress to AIDS. The mechanisms contributing to this control remain incompletely elucidated. In the present study, we have assessed whether cellular host factors could modulate HIV-1 replication post-entry in a controller-discordant couple living with HIV-1.
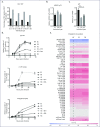
Methods: CD4 T cells from a controller-discordant couple, one partner being an EC and the other an HIV-1 progressor (PR), and healthy controls (HC) were isolated, activated and infected with VSV-G pseudotyped yellow fluorescent protein-encoding single-round HIV-1 virus (HIV-YFP). Viral reverse transcripts, 2-LTR circles and integrated proviral HIV-1 DNA were monitored by quantitative PCR (qPCR) and integration sites were analysed. We further measured LEDGF/p75 and p21 mRNA expression levels by qPCR.
Results: Infection of activated CD4 T cells with HIV-YFP was reduced in EC compared with the PR partner, and HC. Evaluation of viral DNA forms suggested a block after entry and during the early steps of HIV-1 reverse transcription in EC. The integration site distribution pattern in EC, PR and HC was similar. The p21 expression in CD4 T cells of EC was elevated compared with the PR or HC, in line with previous work.
Conclusions: Our study suggests a reduced permissiveness to HIV-1 infection of CD4 T cells from EC due to a block of HIV-1 replication after entry and before integration that might contribute to the EC phenotype in our patient.
Keywords: HIV, elite controllers, HIV eradication, integration.
Figures
References
-
- Blankson JN, Siliciano RF. Elite suppression of HIV-1 replication. Immunity 2008; 29: 845– 847. - PubMed
-
- Deacon NJ, Tsykin A, Solomon A et al. . Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science 1995; 270: 988– 991. - PubMed
-
- Learmont JC, Geczy AF, Mills J et al. . Immunologic and virologic status after 14 to 18 years of infection with an attenuated strain of HIV-1. A report from the Sydney Blood Bank Cohort. N Engl J Med 1999; 340: 1715– 1722. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
