Genetic Diversity, Multidrug Resistance, and Virulence of Citrobacter freundii From Diarrheal Patients and Healthy Individuals
- PMID: 30050870
- PMCID: PMC6052900
- DOI: 10.3389/fcimb.2018.00233
Genetic Diversity, Multidrug Resistance, and Virulence of Citrobacter freundii From Diarrheal Patients and Healthy Individuals
Abstract
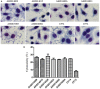
Objectives:Citrobacter freundii is a frequent cause of nosocomial infections and a known cause of diarrheal infections, and has increasingly become multidrug resistant (MDR). In this study, we aimed to determine the genetic diversity, the antimicrobial resistance profiles and in vitro virulence properties of C. freundii from diarrheal patients and healthy individuals. Methods: 82 C. freundii isolates were obtained from human diarrheal outpatients and healthy individuals. Multilocus Sequence Typing (MLST) of seven housekeeping genes was performed. Antimicrobial susceptibility testing was carried out using the disk diffusion method according to the Clinical and Laboratory Standards Institute (CLSI) recommendations. Adhesion and cytotoxicity to HEp-2 cells were assessed. PCR and sequencing were used to identify blaCTX-M, blaSHV, blaTEM, qnrA, qnrB, qnrS, qnrC, qnrD, aac(6')-Ib-cr, and qepA genes. Results: The 82 C. freundii isolates were divided into 76 sequence types (STs) with 65 STs being novel, displaying high genetic diversity. Phylogenetic analysis divided the 82 isolates into 5 clusters. All 82 isolates were sensitive to imipenem (IPM), but resistant to one or more other 16 antibiotics tested. Twenty-six isolates (31.7%) were multidrug resistant to three or more antibiotic classes out of the 10 distinct antibiotic classes tested. Five MDR isolates, all of which were isolated from 2014, harbored one or more of the resistance genes, blaTEM-1, blaCTX-M-9, aac(6')-Ib-cr, qnrS1, qnrB9, and qnrB13. All 11 qnrB-carrying C. freundii isolates belonged to cluster 1, and one C. freundii isolate carried a new qnrB gene (qnrB92). Six isolates showed strong cytotoxicity to HEp-2 cells, one of which was multidrug resistant. Conclusions:C. freundii isolates from human diarrheal outpatients and healthy individuals were diverse with variation in sequence types, antibiotic resistance profiles and virulence properties.
Keywords: Citrobacter freundii; adhesion; cytotoxicity; multidrug resistance; multilocus sequence typing.
Figures

References
-
- Choi S. H., Lee J. E., Park S. J., Kim M. N., Choo E. J., Kwak Y. G., et al. . (2007). Prevalence, microbiology, and clinical characteristics of extended-spectrum beta-lactamase-producing Enterobacter spp., Serratia marcescens, Citrobacter freundii, and Morganella morganii in Korea. Eur. J. Clin. Microbiol. Infect. Dis. 26, 557–561. 10.1007/s10096-007-0308-2 - DOI - PubMed
-
- Clinical and Laboratory Standards Institute (2014). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. CLSI Document M100-S24. Wayne, PA: Clinical and Laboratory Standards Institute.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

