Design tools for MPRA experiments
- PMID: 30052913
- PMCID: PMC6454564
- DOI: 10.1093/bioinformatics/bty150
Design tools for MPRA experiments
Abstract
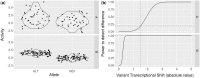
Motivation: Genetic reporter assays are a convenient, relatively inexpensive method for studying the regulation of gene expression. Massively Parallel Reporter Assays (MPRA) are high-throughput functionalization assays that interrogate the transcriptional activity of many genetic variants at once using a library of synthetic barcoded constructs. Despite growing interest in this area, there are few computational tools to design and execute MPRA studies.
Results: We designed an online web-tool and R package that allows for interactive MPRA experimental design encompassing both power analysis and design of constructs. Our tool is tuned using data from real MPRA studies. Users can adjust experimental parameters to examine the predicted effect on assay power as well as upload VCFs for automated construct sequence generation.
Availability and implementation: The MPRA Design Tools web application is available here: https://andrewghazi.shinyapps.io/designmpra/, https://github.com/andrewGhazi/designMPRA and https://github.com/andrewGhazi/mpradesigntools.
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

