Intron-containing algal transgenes mediate efficient recombinant gene expression in the green microalga Chlamydomonas reinhardtii
- PMID: 30053227
- PMCID: PMC6061784
- DOI: 10.1093/nar/gky532
Intron-containing algal transgenes mediate efficient recombinant gene expression in the green microalga Chlamydomonas reinhardtii
Abstract
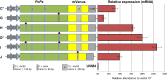
Among green freshwater microalgae, Chlamydomonas reinhardtii has the most comprehensive and developed molecular toolkit, however, advanced genetic and metabolic engineering driven from the nuclear genome is generally hindered by inherently low transgene expression levels. Progressive strain development and synthetic promoters have improved the capacity of transgene expression; however, the responsible regulatory mechanisms are still not fully understood. Here, we elucidate the sequence specific dynamics of native regulatory element insertion into nuclear transgenes. Systematic insertions of the first intron of the ribulose-1,5-bisphosphate carboxylase/oxygenase small subunit 2 (rbcS2i1) throughout codon-optimized coding sequences (CDS) generates optimized algal transgenes which express reliably in C. reinhardtii. The optimal rbcS2i1 insertion site for efficient splicing was systematically determined and improved gene expression rates were shown using a codon-optimized sesquiterpene synthase CDS. Sequential insertions of rbcS2i1 were found to have a step-wise additive effect on all levels of transgene expression, which is likely correlated to a synergy of transcriptional machinery recruitment and mimicking the short average exon lengths natively found in the C. reinhardtii genome. We further demonstrate the value of this optimization with five representative transgene examples and provide guidelines for the design of any desired sequence with this strategy.
Figures



References
-
- Xue J., Niu Y.F., Huang T., Yang W.D., Liu J.S., Li H.Y.. Genetic improvement of the microalga Phaeodactylum tricornutum for boosting neutral lipid accumulation. Metab. Eng. 2015; 27:1–9. - PubMed
-
- Mussgnug J.H. Genetic tools and techniques for Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol. 2015; 99:5407–5418. - PubMed
-
- Scaife M.A., Smith A.G.. Towards developing algal synthetic biology. Biochem. Soc. Trans. 2016; 44:716–722. - PubMed
-
- Jaeger D., Hübner W., Huser T., Mussgnug J.H., Kruse O.. Nuclear transformation and functional gene expression in the oleaginous microalga Monoraphidium neglectum. J. Biotechnol. 2017; 249:10–15. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

