The parts are greater than the whole: the role of semi-infectious particles in influenza A virus biology
- PMID: 30053722
- PMCID: PMC6642613
- DOI: 10.1016/j.coviro.2018.07.002
The parts are greater than the whole: the role of semi-infectious particles in influenza A virus biology
Abstract
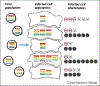
The influenza A virus (IAV) genome is incorporated into newly produced virions through a tightly orchestrated process that is one of the best studied examples of genome packaging by a segmented virus. Despite the remarkable selectivity and efficiency of this process, it is clear that the vast majority of IAV virions are unable to express the full set of essential viral gene products and are thus incapable of productive replication in the absence of complementation. Here, we attempt to reconcile the widespread production of these semi-infectious particles (SIPs) with the high efficiency and selectivity of IAV genome packaging. We also cover what is known and what remains unknown about the consequences of SIP production for the replication and evolution of viral populations.
Copyright © 2018 Elsevier B.V. All rights reserved.
Figures
References
-
- Donald HB, Isaacs A: Counts of influenza virus particles. J Gen Microbiol 1954, 10:457–464. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

