Crystal structure of a novel domain of the motor subunit of the Type I restriction enzyme EcoR124 involved in complex assembly and DNA binding
- PMID: 30054276
- PMCID: PMC6166736
- DOI: 10.1074/jbc.RA118.003978
Crystal structure of a novel domain of the motor subunit of the Type I restriction enzyme EcoR124 involved in complex assembly and DNA binding
Abstract
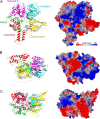
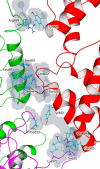
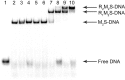
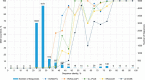
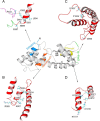
Although EcoR124 is one of the better-studied Type I restriction-modification enzymes, it still presents many challenges to detailed analyses because of its structural and functional complexity and missing structural information. In all available structures of its motor subunit HsdR, responsible for DNA translocation and cleavage, a large part of the HsdR C terminus remains unresolved. The crystal structure of the C terminus of HsdR, obtained with a crystallization chaperone in the form of pHluorin fusion and refined to 2.45 Å, revealed that this part of the protein forms an independent domain with its own hydrophobic core and displays a unique α-helical fold. The full-length HsdR model, based on the WT structure and the C-terminal domain determined here, disclosed a proposed DNA-binding groove lined by positively charged residues. In vivo and in vitro assays with a C-terminal deletion mutant of HsdR supported the idea that this domain is involved in complex assembly and DNA binding. Conserved residues identified through sequence analysis of the C-terminal domain may play a key role in protein-protein and protein-DNA interactions. We conclude that the motor subunit of EcoR124 comprises five structural and functional domains, with the fifth, the C-terminal domain, revealing a unique fold characterized by four conserved motifs in the IC subfamily of Type I restriction-modification systems. In summary, the structural and biochemical results reported here support a model in which the C-terminal domain of the motor subunit HsdR of the endonuclease EcoR124 is involved in complex assembly and DNA binding.
Keywords: C-terminal domain; DNA binding protein; DNA endonuclease; EcoR124; Escherichia coli (E. coli); GFP fusion; HsdR; X-ray crystallography; crystal structure; restriction-modification.
© 2018 Grinkevich et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


References
-
- Manso A. S., Chai M. H., Atack J. M., Furi L., De Ste Croix M., Haigh R., Trappetti C., Ogunniyi A. D., Shewell L. K., Boitano M., Clark T. A., Korlach J., Blades M., Mirkes E., Gorban A. N., et al. (2014) A random six-phase switch regulates pneumococcal virulence via global epigenetic changes. Nat. Commun. 5, 5055 10.1038/ncomms6055 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

