Hippo Signaling Pathway Dysregulation in Human Huntington's Disease Brain and Neuronal Stem Cells
- PMID: 30054496
- PMCID: PMC6063913
- DOI: 10.1038/s41598-018-29319-4
Hippo Signaling Pathway Dysregulation in Human Huntington's Disease Brain and Neuronal Stem Cells
Abstract
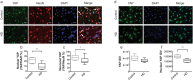
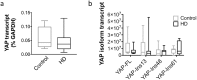
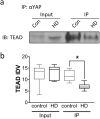
The Hippo signaling pathway is involved in organ size regulation and tumor suppression. Although inhibition of Hippo leads to tumorigenesis, activation of Hippo may play a role in neurodegeneration. Specifically, activation of the upstream regulator, mammalian sterile 20 (STE20)-like kinase 1 (MST1), reduces activity of the transcriptional co-activator Yes-Associated Protein (YAP), thereby mediating oxidative stress-induced neuronal death. Here, we investigated the possible role of this pathway in Huntington's disease (HD) pathogenesis. Our results demonstrate a significant increase in phosphorylated MST1, the active form, in post-mortem HD cortex and in the brains of CAG knock-in HdhQ111/Q111 mice. YAP nuclear localization was also decreased in HD post-mortem cortex and in neuronal stem cells derived from HD patients. Moreover, there was a significant increase in phosphorylated YAP, the inactive form, in HD post-mortem cortex and in HdhQ111/Q111 brain. In addition, YAP was found to interact with huntingtin (Htt) and the chaperone 14-3-3, however this interaction was not altered in the presence of mutant Htt. Lastly, YAP/TEAD interactions and expression of Hippo pathway genes were altered in HD. Together, these results demonstrate that activation of MST1 together with a decrease in nuclear YAP could significantly contribute to transcriptional dysregulation in HD.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell72, 971–983 (1993). - PubMed
-
- MacDonald ME. Huntingtin: alive and well and working in middle management. Sci STKE. 2003;2003:pe48. - PubMed
-
- Glajch KE, Sadri-Vakili G. Epigenetic Mechanisms Involved in Huntington’s Disease Pathogenesis. J Huntingtons Dis. 2015;4:1–15. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

