The MYC transcription factor network: balancing metabolism, proliferation and oncogenesis
- PMID: 30054853
- PMCID: PMC7358075
- DOI: 10.1007/s11684-018-0650-z
The MYC transcription factor network: balancing metabolism, proliferation and oncogenesis
Abstract
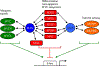
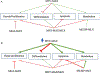
Transcription factor networks have evolved in order to control, coordinate, and separate, the functions of distinct network modules spatially and temporally. In this review we focus on the MYC network (also known as the MAX-MLX Network), a highly conserved super-family of related basic-helix-loop-helix-zipper (bHLHZ) proteins that functions to integrate extracellular and intracellular signals and modulate global gene expression. Importantly the MYC network has been shown to be deeply involved in a broad spectrum of human and other animal cancers. Here we summarize molecular and biological properties of the network modules with emphasis on functional interactions among network members. We suggest that these network interactions serve to modulate growth and metabolism at the transcriptional level in order to balance nutrient demand with supply, to maintain growth homeostasis, and to influence cell fate. Moreover, oncogenic activation of MYC and/or loss of a MYC antagonist, results in an imbalance in the activity of the network as a whole, leading to tumor initiation, progression and maintenance.
Keywords: MAX; MLX; MYC; cancer; network; transcription.
Conflict of interest statement
Compliance with ethics guidelines
Patrick A. Carroll, Brian W. Freie, Haritha Mathsyaraja, and Robert N. Eisenman declare that they have no conflicts of interest. This manuscript is a review article and does not involve a research protocol requiring approval by the relevant institutional review board or ethics committee. Research described from the authors’ laboratory is in full compliance with institutional and national guidelines for the care and use of laboratory animals.
Figures
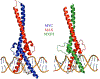


References
-
- Nair SK, Burley SK. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by protooncogenic transcription factors. Cell 2003; 112(2): 193–205 - PubMed
-
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, Otim O, Brown CT, Livi CB, Lee PY, Revilla R, Rust AG, Pan Z, Schilstra MJ, Clarke PJ, Arnone MI, Rowen L, Cameron RA, McClay DR, Hood L, Bolouri H. A genomic regulatory network for development. Science 2002; 295(5560): 1669–1678 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

