Structure and function of the histone chaperone FACT - Resolving FACTual issues
- PMID: 30055319
- PMCID: PMC6349528
- DOI: 10.1016/j.bbagrm.2018.07.008
Structure and function of the histone chaperone FACT - Resolving FACTual issues
Abstract
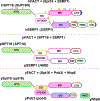
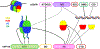
FAcilitates Chromatin Transcription (FACT) has been considered essential for transcription through chromatin mostly based on cell-free experiments. However, FACT inactivation in cells does not cause a significant reduction in transcription. Moreover, not all mammalian cells require FACT for viability. Here we synthesize information from different organisms to reveal the core function(s) of FACT and propose a model that reconciles the cell-free and cell-based observations. We describe FACT structure and nucleosomal interactions, and their roles in FACT-dependent transcription, replication and repair. The variable requirements for FACT among different tumor and non-tumor cells suggest that various FACT-dependent processes have significantly different levels of relative importance in different eukaryotic cells. We propose that the stability of chromatin, which might vary among different cell types, dictates these diverse requirements for FACT to support cell viability. Since tumor cells are among the most sensitive to FACT inhibition, this vulnerability could be exploited for cancer treatment.
Keywords: Cancer; Chromatin; Histone; SPT16; SSRP1; Transcription.
Copyright © 2018. Published by Elsevier B.V.
Figures

Similar articles
-
Complex mutual regulation of facilitates chromatin transcription (FACT) subunits on both mRNA and protein levels in human cells.Cell Cycle. 2013 Aug 1;12(15):2423-34. doi: 10.4161/cc.25452. Epub 2013 Jun 28. Cell Cycle. 2013. PMID: 23839038 Free PMC article.
-
Regulation of chromatin structure and function: insights into the histone chaperone FACT.Cell Cycle. 2021 Mar-Mar;20(5-6):465-479. doi: 10.1080/15384101.2021.1881726. Epub 2021 Feb 16. Cell Cycle. 2021. PMID: 33590780 Free PMC article. Review.
-
The Arabidopsis Histone Chaperone FACT: Role of the HMG-Box Domain of SSRP1.J Mol Biol. 2018 Aug 17;430(17):2747-2759. doi: 10.1016/j.jmb.2018.06.046. Epub 2018 Jun 30. J Mol Biol. 2018. PMID: 29966609
-
The histone chaperone facilitates chromatin transcription (FACT) protein maintains normal replication fork rates.J Biol Chem. 2011 Sep 2;286(35):30504-30512. doi: 10.1074/jbc.M111.264721. Epub 2011 Jul 7. J Biol Chem. 2011. PMID: 21757688 Free PMC article.
-
The FACT Histone Chaperone: Tuning Gene Transcription in the Chromatin Context to Modulate Plant Growth and Development.Front Plant Sci. 2020 Feb 19;11:85. doi: 10.3389/fpls.2020.00085. eCollection 2020. Front Plant Sci. 2020. PMID: 32140163 Free PMC article. Review.
Cited by
-
The Role of PARP1 and PAR in ATP-Independent Nucleosome Reorganisation during the DNA Damage Response.Genes (Basel). 2022 Dec 30;14(1):112. doi: 10.3390/genes14010112. Genes (Basel). 2022. PMID: 36672853 Free PMC article. Review.
-
The HSV-1 ICP22 protein selectively impairs histone repositioning upon Pol II transcription downstream of genes.Nat Commun. 2023 Jul 31;14(1):4591. doi: 10.1038/s41467-023-40217-w. Nat Commun. 2023. PMID: 37524699 Free PMC article.
-
Chromatin Transcription Elongation - A Structural Perspective.J Mol Biol. 2025 Jan 1;437(1):168845. doi: 10.1016/j.jmb.2024.168845. Epub 2024 Oct 29. J Mol Biol. 2025. PMID: 39476950 Review.
-
Evidence that dissociation of Spt16 from transcribed genes is partially dependent on RNA Polymerase II termination.Transcription. 2019 Aug-Oct;10(4-5):195-206. doi: 10.1080/21541264.2019.1685837. Epub 2019 Dec 6. Transcription. 2019. PMID: 31809228 Free PMC article.
-
Structural Transition of the Nucleosome during Transcription Elongation.Cells. 2023 May 14;12(10):1388. doi: 10.3390/cells12101388. Cells. 2023. PMID: 37408222 Free PMC article. Review.
References
-
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ, Crystal structure of the nucleosome core particle at 2.8 A resolution, Nature, 389 (1997) 251–260. - PubMed
-
- Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ, Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 A resolution, J Mol Biol, 319 (2002) 1097–1113. - PubMed
-
- Andrews AJ, Luger K, Nucleosome structure(s) and stability: variations on a theme, Annu Rev Biophys, 40 (2011) 99–117. - PubMed
-
- Kireeva ML, Walter W, Tchernajenko V, Bondarenko V, Kashlev M, Studitsky VM, Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription, Mol Cell, 9 (2002) 541–552. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

