Exposing the Elusive Exocyst Structure
- PMID: 30055895
- PMCID: PMC6108956
- DOI: 10.1016/j.tibs.2018.06.012
Exposing the Elusive Exocyst Structure
Abstract
A major challenge for a molecular understanding of membrane trafficking has been the elucidation of high-resolution structures of large, multisubunit tethering complexes that spatially and temporally control intracellular membrane fusion. Exocyst is a large hetero-octameric protein complex proposed to tether secretory vesicles at the plasma membrane to provide quality control of soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE)-mediated membrane fusion. Breakthroughs in methodologies, including sample preparation, biochemical characterization, fluorescence microscopy, and single-particle cryoelectron microscopy, are providing critical insights into the structure and function of the exocyst. These studies now pose more questions than answers for understanding fundamental functional mechanisms, and they open wide the door for future studies to elucidate interactions with protein and membrane partners, potential conformational changes, and molecular insights into tethering reactions.
Keywords: Rab GTPase; Rho GTPase; SNARE; exocyst; exocytosis; membrane fusion; membrane trafficking; tethering complex.
Copyright © 2018 Elsevier Ltd. All rights reserved.
Figures
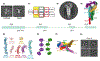
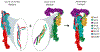
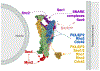
References
-
- Pfeffer SR (1999) Transport-vesicle targeting: tethers before SNAREs. Nat Cell Biol 1 (1), E17–22. - PubMed
-
- Yu IM and Hughson FM (2010) Tethering factors as organizers of intracellular vesicular traffic. Annu Rev Cell Dev Biol 26, 137–56. - PubMed
-
- Gillingham AK and Munro S (2016) Finding the Golgi: Golgin Coiled-Coil Proteins Show the Way. Trends Cell Biol 26 (6), 399–408. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

