SUMO-defective c-Maf preferentially transactivates Il21 to exacerbate autoimmune diabetes
- PMID: 30059018
- PMCID: PMC6118635
- DOI: 10.1172/JCI98786
SUMO-defective c-Maf preferentially transactivates Il21 to exacerbate autoimmune diabetes
Abstract
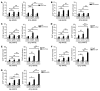
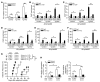
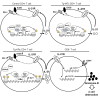
SUMOylation is involved in the development of several inflammatory diseases, but the physiological significance of SUMO-modulated c-Maf in autoimmune diabetes is not completely understood. Here, we report that an age-dependent attenuation of c-Maf SUMOylation in CD4+ T cells is positively correlated with the IL-21-mediated diabetogenesis in NOD mice. Using 2 strains of T cell-specific transgenic NOD mice overexpressing wild-type c-Maf (Tg-WTc) or SUMOylation site-mutated c-Maf (Tg-KRc), we demonstrated that Tg-KRc mice developed diabetes more rapidly than Tg-WTc mice in a CD4+ T cell-autonomous manner. Moreover, SUMO-defective c-Maf preferentially transactivated Il21 to promote the development of CD4+ T cells with an extrafollicular helper T cell phenotype and expand the numbers of granzyme B-producing effector/memory CD8+ T cells. Furthermore, SUMO-defective c-Maf selectively inhibited recruitment of Daxx/HDAC2 to the Il21 promoter and enhanced histone acetylation mediated by CREB-binding protein (CBP) and p300. Using pharmacological interference with CBP/p300, we illustrated that CBP30 treatment ameliorated c-Maf-mediated/IL-21-based diabetogenesis. Taken together, our results show that the SUMOylation status of c-Maf has a stronger regulatory effect on IL-21 than the level of c-Maf expression, through an epigenetic mechanism. These findings provide new insights into how SUMOylation modulates the pathogenesis of autoimmune diabetes in a T cell-restricted manner and on the basis of a single transcription factor.
Keywords: Autoimmunity; Cytokines; Epigenetics; Immunology; T cells.
Conflict of interest statement
Figures





References
-
- Decque A, et al. Sumoylation coordinates the repression of inflammatory and anti-viral gene-expression programs during innate sensing. Nat Immunol. 2016;17(2):140–149. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

