DNA replication stress triggers rapid DNA replication fork breakage by Artemis and XPF
- PMID: 30059501
- PMCID: PMC6085069
- DOI: 10.1371/journal.pgen.1007541
DNA replication stress triggers rapid DNA replication fork breakage by Artemis and XPF
Abstract
DNA replication stress (DRS) leads to the accumulation of stalled DNA replication forks leaving a fraction of genomic loci incompletely replicated, a source of chromosomal rearrangements during their partition in mitosis. MUS81 is known to limit the occurrence of chromosomal instability by processing these unresolved loci during mitosis. Here, we unveil that the endonucleases ARTEMIS and XPF-ERCC1 can also induce stalled DNA replication forks cleavage through non-epistatic pathways all along S and G2 phases of the cell cycle. We also showed that both nucleases are recruited to chromatin to promote replication fork restart. Finally, we found that rapid chromosomal breakage controlled by ARTEMIS and XPF is important to prevent mitotic segregation defects. Collectively, these results reveal that Rapid Replication Fork Breakage (RRFB) mediated by ARTEMIS and XPF in response to DRS contributes to DNA replication efficiency and limit chromosomal instability.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
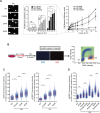
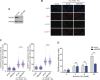

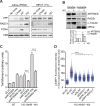
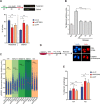
References
-
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37(4):492–502. Epub 2010/03/02. 10.1016/j.molcel.2010.01.021 ; PubMed Central PMCID: PMC2958316. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

