Hierarchical multiscale Bayesian algorithm for robust MEG/EEG source reconstruction
- PMID: 30059734
- PMCID: PMC6214686
- DOI: 10.1016/j.neuroimage.2018.07.056
Hierarchical multiscale Bayesian algorithm for robust MEG/EEG source reconstruction
Abstract
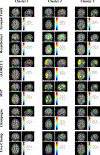
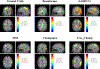
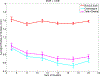
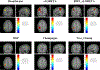
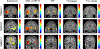
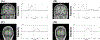
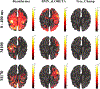
In this paper, we present a novel hierarchical multiscale Bayesian algorithm for electromagnetic brain imaging using magnetoencephalography (MEG) and electroencephalography (EEG). In particular, we present a solution to the source reconstruction problem for sources that vary in spatial extent. We define sensor data measurements using a generative probabilistic graphical model that is hierarchical across spatial scales of brain regions and voxels. We then derive a novel Bayesian algorithm for probabilistic inference with this graphical model. This algorithm enables robust reconstruction of sources that have different spatial extent, from spatially contiguous clusters of dipoles to isolated dipolar sources. We compare the new algorithm with several representative benchmarks on both simulated and real brain activities. The source locations and the correct estimation of source time courses used for the simulated data are chosen to test the performance on challenging source configurations. In simulations, performance of the novel algorithm shows superiority to several existing benchmark algorithms. We also demonstrate that the new algorithm is more robust to correlated brain activity present in real MEG and EEG data and is able to resolve distinct and functionally relevant brain areas with real MEG and EEG datasets.
Keywords: Bayesian; Brain mapping; Electroencephalography; Magnetoencephalography.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Figures








References
-
- Sekihara K and Nagarajan SS. Adaptive spatial filters for electromagnetic brain imaging. Springer-Verlag, Berlin, Heidelber, 2008.
-
- Sekihara Kensuke and Nagarajan Srikatan S. Electromagnetic brain imaging: A Bayesian perspective. Springer-Verlag, Berlin, Heidelber, 2015.
-
- Wu Wei, Nagarajan Srikantan, and Chen Zhe. Bayesian machine learning: EEG/MEG signal processing measurements. IEEE Signal Processing Magazine, 33(1):14–36, 2016.
-
- H¨am¨al¨ainen MS and Ilmoniemi RJ. Interpreting measured magnetic fields of the brain: Estimates of current distributions Technical Report TKK-F-A559, Helsinki University of Technology, 1984.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

