Histone methyltransferase Smyd1 regulates mitochondrial energetics in the heart
- PMID: 30061404
- PMCID: PMC6099878
- DOI: 10.1073/pnas.1800680115
Histone methyltransferase Smyd1 regulates mitochondrial energetics in the heart
Abstract
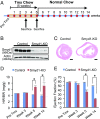
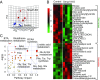
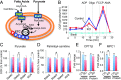
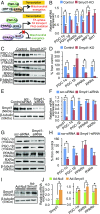
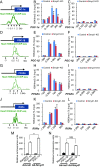
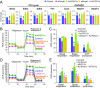
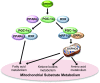
Smyd1, a muscle-specific histone methyltransferase, has established roles in skeletal and cardiac muscle development, but its role in the adult heart remains poorly understood. Our prior work demonstrated that cardiac-specific deletion of Smyd1 in adult mice (Smyd1-KO) leads to hypertrophy and heart failure. Here we show that down-regulation of mitochondrial energetics is an early event in these Smyd1-KO mice preceding the onset of structural abnormalities. This early impairment of mitochondrial energetics in Smyd1-KO mice is associated with a significant reduction in gene and protein expression of PGC-1α, PPARα, and RXRα, the master regulators of cardiac energetics. The effect of Smyd1 on PGC-1α was recapitulated in primary cultured rat ventricular myocytes, in which acute siRNA-mediated silencing of Smyd1 resulted in a greater than twofold decrease in PGC-1α expression without affecting that of PPARα or RXRα. In addition, enrichment of histone H3 lysine 4 trimethylation (a mark of gene activation) at the PGC-1α locus was markedly reduced in Smyd1-KO mice, and Smyd1-induced transcriptional activation of PGC-1α was confirmed by luciferase reporter assays. Functional confirmation of Smyd1's involvement showed an increase in mitochondrial respiration capacity induced by overexpression of Smyd1, which was abolished by siRNA-mediated PGC-1α knockdown. Conversely, overexpression of PGC-1α rescued transcript expression and mitochondrial respiration caused by silencing Smyd1 in cardiomyocytes. These findings provide functional evidence for a role of Smyd1, or any member of the Smyd family, in regulating cardiac energetics in the adult heart, which is mediated, at least in part, via modulating PGC-1α.
Keywords: PGC-1a; Smyd1; heart; metabolism; systems biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Wu J, et al. Biochemical characterization of human SET and MYND domain-containing protein 2 methyltransferase. Biochemistry. 2011;50:6488–6497. - PubMed
-
- Gottlieb PD, et al. Bop encodes a muscle-restricted protein containing MYND and SET domains and is essential for cardiac differentiation and morphogenesis. Nat Genet. 2002;31:25–32. - PubMed
-
- Borlak J, Thum T. Hallmarks of ion channel gene expression in end-stage heart failure. FASEB J. 2003;17:1592–1608. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

